
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CAB39-RHBDD1 (FusionGDB2 ID:HG51719TG84236) |
Fusion Gene Summary for CAB39-RHBDD1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CAB39-RHBDD1 | Fusion gene ID: hg51719tg84236 | Hgene | Tgene | Gene symbol | CAB39 | RHBDD1 | Gene ID | 51719 | 84236 |
| Gene name | calcium binding protein 39 | rhomboid domain containing 1 | |
| Synonyms | CGI-66|MO25 | RHBDL4|RRP4 | |
| Cytomap | ('CAB39')('RHBDD1') 2q37.1 | 2q36.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium-binding protein 39MO25alpha | rhomboid-related protein 4rhomboid domain-containing protein 1rhomboid-like protein 4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000258418, ENST00000409788, ENST00000410084, ENST00000484398, | ||
| Fusion gene scores | * DoF score | 7 X 4 X 5=140 | 8 X 7 X 4=224 |
| # samples | 9 | 8 | |
| ** MAII score | log2(9/140*10)=-0.637429920615292 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/224*10)=-1.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CAB39 [Title/Abstract] AND RHBDD1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CAB39(231624830)-RHBDD1(227860147), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CAB39-RHBDD1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CAB39-RHBDD1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CAB39-RHBDD1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CAB39-RHBDD1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CAB39-RHBDD1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CAB39 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | 24393035 |
| Hgene | CAB39 | GO:0018105 | peptidyl-serine phosphorylation | 24393035 |
| Hgene | CAB39 | GO:0023014 | signal transduction by protein phosphorylation | 24393035 |
| Hgene | CAB39 | GO:0035556 | intracellular signal transduction | 24393035 |
| Hgene | CAB39 | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 19892943|24393035 |
 Fusion gene breakpoints across CAB39 (5'-gene) Fusion gene breakpoints across CAB39 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
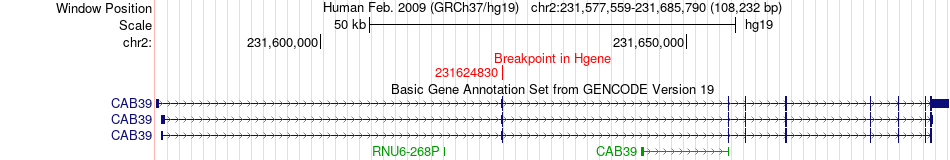 |
 Fusion gene breakpoints across RHBDD1 (3'-gene) Fusion gene breakpoints across RHBDD1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
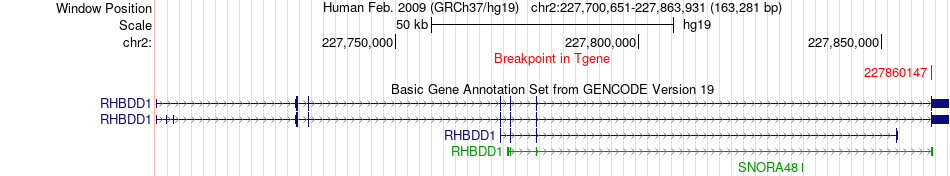 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A8BX-01A | CAB39 | chr2 | 231624830 | - | RHBDD1 | chr2 | 227860147 | + |
| ChimerDB4 | SARC | TCGA-DX-A8BX-01A | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
Top |
Fusion Gene ORF analysis for CAB39-RHBDD1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000258418 | ENST00000493526 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| 5CDS-3UTR | ENST00000409788 | ENST00000493526 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| 5CDS-3UTR | ENST00000410084 | ENST00000493526 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| 5CDS-intron | ENST00000258418 | ENST00000409053 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| 5CDS-intron | ENST00000409788 | ENST00000409053 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| 5CDS-intron | ENST00000410084 | ENST00000409053 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| Frame-shift | ENST00000258418 | ENST00000392062 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| Frame-shift | ENST00000409788 | ENST00000392062 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| Frame-shift | ENST00000410084 | ENST00000392062 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| In-frame | ENST00000258418 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| In-frame | ENST00000409788 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| In-frame | ENST00000410084 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| intron-3CDS | ENST00000484398 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| intron-3CDS | ENST00000484398 | ENST00000392062 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| intron-3UTR | ENST00000484398 | ENST00000493526 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
| intron-intron | ENST00000484398 | ENST00000409053 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000258418 | CAB39 | chr2 | 231624830 | + | ENST00000341329 | RHBDD1 | chr2 | 227860147 | + | 4328 | 543 | 430 | 2 | 143 |
| ENST00000410084 | CAB39 | chr2 | 231624830 | + | ENST00000341329 | RHBDD1 | chr2 | 227860147 | + | 4197 | 412 | 738 | 1100 | 120 |
| ENST00000409788 | CAB39 | chr2 | 231624830 | + | ENST00000341329 | RHBDD1 | chr2 | 227860147 | + | 4453 | 668 | 715 | 185 | 176 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000258418 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + | 0.78202206 | 0.21797797 |
| ENST00000410084 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + | 0.7958127 | 0.2041873 |
| ENST00000409788 | ENST00000341329 | CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860147 | + | 0.808517 | 0.19148298 |
Top |
Fusion Genomic Features for CAB39-RHBDD1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860146 | + | 1.41E-08 | 1 |
| CAB39 | chr2 | 231624830 | + | RHBDD1 | chr2 | 227860146 | + | 1.41E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
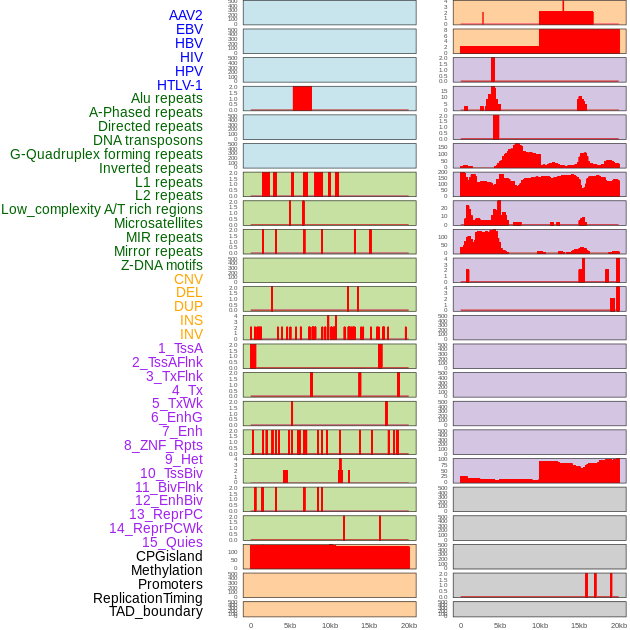 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
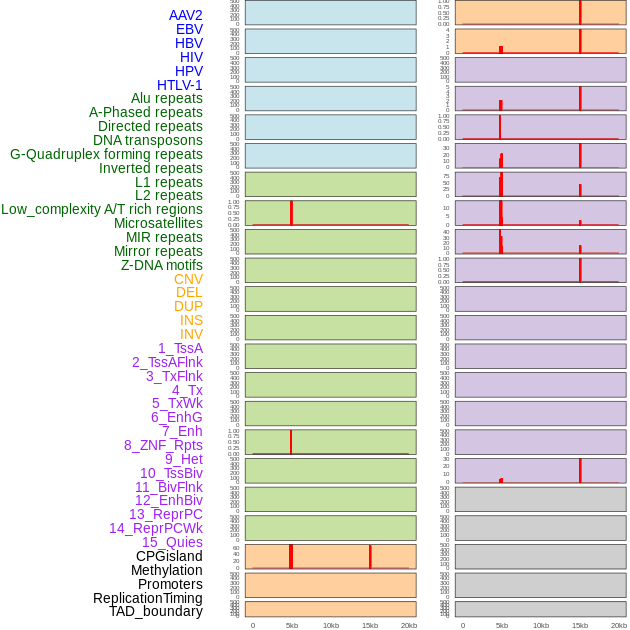 |
Top |
Fusion Protein Features for CAB39-RHBDD1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:231624830/chr2:227860147) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 301_315 | 285 | 316.0 | Region | VCP/p97-interacting motif (VIM) | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 301_315 | 285 | 316.0 | Region | VCP/p97-interacting motif (VIM) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 269_284 | 285 | 316.0 | Region | Ubiquitin-binding domain (UBD) | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 269_284 | 285 | 316.0 | Region | Ubiquitin-binding domain (UBD) | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 128_138 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 158_180 | 285 | 316.0 | Topological domain | Extracellular | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 1_21 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 202_315 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 43_106 | 285 | 316.0 | Topological domain | Extracellular | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 128_138 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 158_180 | 285 | 316.0 | Topological domain | Extracellular | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 1_21 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 202_315 | 285 | 316.0 | Topological domain | Cytoplasmic | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 43_106 | 285 | 316.0 | Topological domain | Extracellular | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 107_127 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 139_157 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 181_201 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000341329 | 5 | 7 | 22_42 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 107_127 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 139_157 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 181_201 | 285 | 316.0 | Transmembrane | Helical | |
| Tgene | RHBDD1 | chr2:231624830 | chr2:227860147 | ENST00000392062 | 7 | 9 | 22_42 | 285 | 316.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for CAB39-RHBDD1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >12223_12223_1_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000258418_RHBDD1_chr2_227860147_ENST00000341329_length(transcript)=4328nt_BP=543nt GTGCGCACGTCAAAGCCGGGCTCGGGCCGCAAGCGGGGCGAGGGGTTCGGGGAGCGGCGCGGCCTGGGAGACACAGAGCCTTCAGGCGCC GGGGCGGGGGCACAGGCGAAGACTAAGGCGGCGCCGGGCCCCACAGCAGCAGCCGCAGCCCAAGCGAGCGCAGCAGCGCGGCGGCAGCCG CGGGAGCCCCTGGGCAGCCGTCCGCCCGCGCAGCCGCCGCCGCCGCGGGAGCCCGTCGCCGGGAGCAGGAGCGGGCGGAAGACAACGGAG GGGCCGAGCGTCCGAGCCACTCCGCGGGGACCGAACGAGCAGCCCGAAGCGGCGGCGGCCGAGGACGGGGACAGCGACGACGCGGAGGCA GAGAAGGGAACGCCCGGCCCAGCCCCGTAGCACAGGCGGAGTGCAGCGGAGGCCCCTGCCGCTGCCGTCATGCCGTTCCCGTTTGGGAAG TCTCACAAATCTCCAGCAGACATTGTGAAGAATCTGAAGGAGAGCATGGCTGTTCTGGAAAAGCAAGACATTTCTGATAAAAAAGCAGAA AAGGAAATACCAGAAATAGCCCACCACCCTACGGGTTTCATCTCTCACCAGAAGAAATGAGGAGACAGCGGCTTCACAGATTCGATAGCC AGTGAGGTGGCATCTTGGGAAGACATGGCCTATTCGTGTAATTATTGCCCATTTGGCTCATTCCCCAAGCCCCTAATTCATTTTAATTCA TTTTAAACAAAAGCAGAGTACACCGGTATTGCTCCAGATCGCTCACATCACCTGGGACAGTCCCATGGCCCCTATGAGTCAACTCACAGC TTGCGGGGAGTGGGCCTTCTCCTGGCCTTGTTCTTGCTCATAAACAGGTCACTTCCTCCATGAAGAGACCAGTTTCCACGCTCCCATCTC TCACTGCTGACTCAGCGATGCCTCTGCCTCGGTCTGCTTTTGAAGACTGTGACCTTCACCAGGAGGTTTTACTTACACCAGTCGGGAAGA TTAGTCCCTCATTCTGCCTGGAGTGCCCCGTGTTTGACTTGGCAGCGGGTGTGGAGCCATCCCCGCGTCCTCCTGGCGCATTGCCACTGT GGCTGTCCAGGAACAGGATGTGGCTGCCTTGGCCGAATGTTGTCCTACTCTCCCCAACCCCGGCGCCTCAGCTCCTCAGCTCCTCGGGCC CCTGCGTCTGGCTGGTGTTTGCAGGGCTTTCGCTCTGCTCTGGTATTGCTCTGCCTTTATAGAAAGTCTTATTGAAGAAGTGTAAGAAAG ACCTAAGGTGGGGAAGACTCCTACACACACCATTAGTATCAGTGACACCAGCAATGTAGGTTCCCAGCCCCTTCCCAGTGGCAGCTTGTG TGTCCAGGAGATAGGACATCATTTAACGCATCAGCAAAGTAGCAGCAGATGCCACATACAGAGTAGAGCGAAGGCATTTGGTGGATCGGT CACTAGAGATCTATCTTGCAGAAAGTATGTTTTTCCTCATAAAAGTGCCTCTTAATTGGCCATTGTACCAGCCACTTGTCCTAGCCAAAT GTCCAAAACACGCCCTTGGGCCCCGCCACGTTACAATCCACAGATTGTCTGTCTGAGTCGTTTAAGGCATTTCCTGGTGCTTGTGTTCCA TGAATAAAAGGACAAAGTCAGAAGATCACTGATGTCTTACTGTCAACAGAGATATTTTAAAAGAGAGAAGCAGGAAAAGATCTTCCTTTT TTGATCTACAACTTATATAGTTTTCTGATTATGCACATAATAGATATGCCTTCCAGATGCATAAGGCAAACATCTGGAAAGAAATATACC CAAATCTTAGCAGGGGTTATCTTTGGGAGTGGAGTACATGGGATTTTGCTTTCTTCATTTTTATAATTTTATATTACTGTCTTGGAAGAT GTGTTTATGTGTGTGTGTTACTTTTACAATCAGGAAAACATATTTAATAACATATAGTCAAGAAAACAGACTTAAAAATAAATACTATGT GTCCATTGAGAAAATTCACAATATAAACAGAAATACAAATAAATACATACACAATTTTAAAGTCACCTGTAGCCCTACCCTTAGAGGTAC CCAGGGTTAACATTTTGGTGGTATTGTCTTATCAATTTTTCCGTTGATACATTCAGCAAATTTGGAGCACATTGACCATGGAGTTTTGTG TCCAAATCCAATCTGAATTTACCTGGAAGAGGCCTTGACACCTGCATGGAAATGAGCTAAGAAAACCACTGGAGCCTTGGGAGCTCTTTG GCCTCCTGGCTGGCCCAGTAATATCTGAGCTCCTTTGGTTAATTTATAACTGATATAAAACTACATCTTCTTTATAATATAAATTGTACC TGTGAGTCTAGAAGCTTTAAATGTGTTTAAATTAAAATATTCAAGCTAAATGTTACTGCTCTCTCCCAAATTCTGTAAGTTTGACTCCCG TTACCCCAATTAGAAGTAACTTCTTTGTTTCATGCCACTTTTATAGCATTTGGTAATTCTGCTATAACACATCTTGCCCCTATTATTAAC TGTGCACAGCTACACAAAGGTGTGCCTTCTACGTGGGAACATGGATTGTGAATGACTCTGTAATGAGGCCTGAGTCTTAGTTATCTTTCC ACTCACTCCCCGTCTCCCCTTTCCAACCCCAAAGGCTCACGATAGGGGCTCACTAAATGTCAGTGTTTCACCAAAGTATTTTTTCCATTG TATTAAGAGTCCAGTCACTGTATATGGAAGTATTTTATTTTTTATTTTTTTATATCACTTGAGTCCACTAGTAGTACTTCCTTGCTCTGT TTGACTTGTCAGATACAAAGACACGGGATTAGATTTTGGGTGGTAAAATTGTGATACGCATGGCTGTTGATGGAGTGGAACATCTTAGTG ATGTGAGAAAGGTCATTTTAGTTATAAATGTAAACCAATTACTTTAGCACAACAATAAAGATGTTCTGGAAATTATTATAATTATTTTAG TTAATGGACTTGCCTGTAACAAGTGCTTATGTTTGGAATGTCCATTTTATTTTATTTTCCTCTACAATAAGGAATCCCTAAAGTTATGCC AACAAACTGGAATTGGATTGAGAAAGACAAAAGCAAGTGAGCAAGTGTATTAGACTAATTATCTCTAGTAGGGAATAAGCACTTAGAGCT AATTTTTTAATTTGGTTTATTTATTAGTAGCTCCCTGATTGTCTGTTAAGCCCCTGGTTCCCACAGTTTGGTGTGGTCAGTCAACTTAAG CGACCTTTTAATGCAGAATCTTTACTTGGTTCTGAAGAGGAGGCAGTAGTTAAAATTGCTTCCTCTTAATGACTCTGAATGTATTGCCTG TGATTCCAAAGTTGTACATAATTGTTCAGACCTCCTCCATCCTTTTAAATTGCCTGCTGCAGTAAATAACTAGTTTGAGTAGAACTAGAT CCTGTCTATCTATTTGGCACATGTTCTGCTGCCTGGGGAGTAAGCAAGCTAAAGGGATGAGAAAGACCACCTCCCCCTACCCTGGAAATT GCACTGCAAGGCAGGGCGAGAATGGGGTAGCTGGCAGACCTGGCCTCCTTGTTCCCAGTCTTAGTTTTTCTTGAGAGATTCAGTATTCAG TAAAGAATAGCATTCAATTAGTCAAAAAATATATATATAACTTCTTCCTTTCCCTTCCCATGAATCATTGCAGTCATTCCCTAAGTTTCT TCTCTTTTTTTTTCATGGCTGCTAGTATTTTATTTTAGAATCACAGACTCTCAGCTTAGAGAGTCACAGTATCTCAATTATTTGCTCTGT TCAATACTTAACAATACCTCCTCACTCAATTCTACATAACTCCAGCTTAGTCTTCCAAAATTCACTTTCATGATGCCACTCAGCATCTCA AATACCTTTCATTGGCTCTCTGCTGCCAAAGGATAAAGGTCAAAGTCATTAGCCTCAACAGTGGGCTTCAACCAGCCTTTGGACCTCAGC CCCATTTATCCATCACAGAGGCTGGTAACTAGTCTCACTGCTCAGGCTGTGAGTGTTCCTGATCTTGTGACATTCTGTGCTGTGCTTTTA CATGGAACAGCTCTTTCCTCTCTCTTGGCCCATTCAGATCCTCCTCATCCAGCCCCCATCTGAATCCTGTAACAGACACAACCACATCTA CACAGTTTCCACATACCACTTGGAATTGCTGGTTATGCTGCAATGAGAGAATGTCTTTGTCTTTAAATTGTAAGCTTCATGAGGGCAGGG ACCATCTGTTTCTCTGTCTCTCCTCACAGTGCCTTGAGCAGTACGTTGTCTGGAATGTCAGAACCAATAAATACTTGTGGATTGAAACAA AAAGAGAA >12223_12223_1_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000258418_RHBDD1_chr2_227860147_ENST00000341329_length(amino acids)=143AA_BP= MTAAAGASAALRLCYGAGPGVPFSASASSLSPSSAAAASGCSFGPRGVARTLGPSVVFRPLLLPATGSRGGGGCAGGRLPRGSRGCRRAA ALAWAAAAAVGPGAALVFACAPAPAPEGSVSPRPRRSPNPSPRLRPEPGFDVR -------------------------------------------------------------- >12223_12223_2_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000409788_RHBDD1_chr2_227860147_ENST00000341329_length(transcript)=4453nt_BP=668nt AGTCGGGCGGGCGGGGAAGTGCGGGCCGGGTCAACTGTCACCGGCTGCACCCGGGCTTCAGCGCCTCGCTCCCGCCCCTGGTAAACTTCC TGTTGTCGGGAGGCAGATACCAGCTTTGTGACCAATTCAGGAAAATGCCTAACTGATTTCTGGTTAGGAATTGGGCGGTCGGAGCGTTCT GGAGTTCAATGGAACCACCCCAAGCGCAGACTGCGTGCAGGCGCCGTAGTCGGCCCGCACCGGAGGGATTCAAAGGTGAAGCGGGCCCCG GTCCCCTCCCCCGAGGAGCTTAGGCTCTGGTGTGAGAGATAAGACGAGAACACAAATAACTGCGATACCGGGAAGGAGGTGATAAGTCCT GTGAAAGAACGACGGAGTGCTAGAACGGCACCCGGAAAAACCGAACCGATCACTTCTGCCTCGCAGAAACCTGGAAGGCTTCCTGGGCGG GGGATGCTGAGCAGAGTCTTGAAGGCCTGATGGGATTTAGGAGAGGAGGATTGCAGAAGAGGTAGCACAGGCGGAGTGCAGCGGAGGCCC CTGCCGCTGCCGTCATGCCGTTCCCGTTTGGGAAGTCTCACAAATCTCCAGCAGACATTGTGAAGAATCTGAAGGAGAGCATGGCTGTTC TGGAAAAGCAAGACATTTCTGATAAAAAAGCAGAAAAGGAAATACCAGAAATAGCCCACCACCCTACGGGTTTCATCTCTCACCAGAAGA AATGAGGAGACAGCGGCTTCACAGATTCGATAGCCAGTGAGGTGGCATCTTGGGAAGACATGGCCTATTCGTGTAATTATTGCCCATTTG GCTCATTCCCCAAGCCCCTAATTCATTTTAATTCATTTTAAACAAAAGCAGAGTACACCGGTATTGCTCCAGATCGCTCACATCACCTGG GACAGTCCCATGGCCCCTATGAGTCAACTCACAGCTTGCGGGGAGTGGGCCTTCTCCTGGCCTTGTTCTTGCTCATAAACAGGTCACTTC CTCCATGAAGAGACCAGTTTCCACGCTCCCATCTCTCACTGCTGACTCAGCGATGCCTCTGCCTCGGTCTGCTTTTGAAGACTGTGACCT TCACCAGGAGGTTTTACTTACACCAGTCGGGAAGATTAGTCCCTCATTCTGCCTGGAGTGCCCCGTGTTTGACTTGGCAGCGGGTGTGGA GCCATCCCCGCGTCCTCCTGGCGCATTGCCACTGTGGCTGTCCAGGAACAGGATGTGGCTGCCTTGGCCGAATGTTGTCCTACTCTCCCC AACCCCGGCGCCTCAGCTCCTCAGCTCCTCGGGCCCCTGCGTCTGGCTGGTGTTTGCAGGGCTTTCGCTCTGCTCTGGTATTGCTCTGCC TTTATAGAAAGTCTTATTGAAGAAGTGTAAGAAAGACCTAAGGTGGGGAAGACTCCTACACACACCATTAGTATCAGTGACACCAGCAAT GTAGGTTCCCAGCCCCTTCCCAGTGGCAGCTTGTGTGTCCAGGAGATAGGACATCATTTAACGCATCAGCAAAGTAGCAGCAGATGCCAC ATACAGAGTAGAGCGAAGGCATTTGGTGGATCGGTCACTAGAGATCTATCTTGCAGAAAGTATGTTTTTCCTCATAAAAGTGCCTCTTAA TTGGCCATTGTACCAGCCACTTGTCCTAGCCAAATGTCCAAAACACGCCCTTGGGCCCCGCCACGTTACAATCCACAGATTGTCTGTCTG AGTCGTTTAAGGCATTTCCTGGTGCTTGTGTTCCATGAATAAAAGGACAAAGTCAGAAGATCACTGATGTCTTACTGTCAACAGAGATAT TTTAAAAGAGAGAAGCAGGAAAAGATCTTCCTTTTTTGATCTACAACTTATATAGTTTTCTGATTATGCACATAATAGATATGCCTTCCA GATGCATAAGGCAAACATCTGGAAAGAAATATACCCAAATCTTAGCAGGGGTTATCTTTGGGAGTGGAGTACATGGGATTTTGCTTTCTT CATTTTTATAATTTTATATTACTGTCTTGGAAGATGTGTTTATGTGTGTGTGTTACTTTTACAATCAGGAAAACATATTTAATAACATAT AGTCAAGAAAACAGACTTAAAAATAAATACTATGTGTCCATTGAGAAAATTCACAATATAAACAGAAATACAAATAAATACATACACAAT TTTAAAGTCACCTGTAGCCCTACCCTTAGAGGTACCCAGGGTTAACATTTTGGTGGTATTGTCTTATCAATTTTTCCGTTGATACATTCA GCAAATTTGGAGCACATTGACCATGGAGTTTTGTGTCCAAATCCAATCTGAATTTACCTGGAAGAGGCCTTGACACCTGCATGGAAATGA GCTAAGAAAACCACTGGAGCCTTGGGAGCTCTTTGGCCTCCTGGCTGGCCCAGTAATATCTGAGCTCCTTTGGTTAATTTATAACTGATA TAAAACTACATCTTCTTTATAATATAAATTGTACCTGTGAGTCTAGAAGCTTTAAATGTGTTTAAATTAAAATATTCAAGCTAAATGTTA CTGCTCTCTCCCAAATTCTGTAAGTTTGACTCCCGTTACCCCAATTAGAAGTAACTTCTTTGTTTCATGCCACTTTTATAGCATTTGGTA ATTCTGCTATAACACATCTTGCCCCTATTATTAACTGTGCACAGCTACACAAAGGTGTGCCTTCTACGTGGGAACATGGATTGTGAATGA CTCTGTAATGAGGCCTGAGTCTTAGTTATCTTTCCACTCACTCCCCGTCTCCCCTTTCCAACCCCAAAGGCTCACGATAGGGGCTCACTA AATGTCAGTGTTTCACCAAAGTATTTTTTCCATTGTATTAAGAGTCCAGTCACTGTATATGGAAGTATTTTATTTTTTATTTTTTTATAT CACTTGAGTCCACTAGTAGTACTTCCTTGCTCTGTTTGACTTGTCAGATACAAAGACACGGGATTAGATTTTGGGTGGTAAAATTGTGAT ACGCATGGCTGTTGATGGAGTGGAACATCTTAGTGATGTGAGAAAGGTCATTTTAGTTATAAATGTAAACCAATTACTTTAGCACAACAA TAAAGATGTTCTGGAAATTATTATAATTATTTTAGTTAATGGACTTGCCTGTAACAAGTGCTTATGTTTGGAATGTCCATTTTATTTTAT TTTCCTCTACAATAAGGAATCCCTAAAGTTATGCCAACAAACTGGAATTGGATTGAGAAAGACAAAAGCAAGTGAGCAAGTGTATTAGAC TAATTATCTCTAGTAGGGAATAAGCACTTAGAGCTAATTTTTTAATTTGGTTTATTTATTAGTAGCTCCCTGATTGTCTGTTAAGCCCCT GGTTCCCACAGTTTGGTGTGGTCAGTCAACTTAAGCGACCTTTTAATGCAGAATCTTTACTTGGTTCTGAAGAGGAGGCAGTAGTTAAAA TTGCTTCCTCTTAATGACTCTGAATGTATTGCCTGTGATTCCAAAGTTGTACATAATTGTTCAGACCTCCTCCATCCTTTTAAATTGCCT GCTGCAGTAAATAACTAGTTTGAGTAGAACTAGATCCTGTCTATCTATTTGGCACATGTTCTGCTGCCTGGGGAGTAAGCAAGCTAAAGG GATGAGAAAGACCACCTCCCCCTACCCTGGAAATTGCACTGCAAGGCAGGGCGAGAATGGGGTAGCTGGCAGACCTGGCCTCCTTGTTCC CAGTCTTAGTTTTTCTTGAGAGATTCAGTATTCAGTAAAGAATAGCATTCAATTAGTCAAAAAATATATATATAACTTCTTCCTTTCCCT TCCCATGAATCATTGCAGTCATTCCCTAAGTTTCTTCTCTTTTTTTTTCATGGCTGCTAGTATTTTATTTTAGAATCACAGACTCTCAGC TTAGAGAGTCACAGTATCTCAATTATTTGCTCTGTTCAATACTTAACAATACCTCCTCACTCAATTCTACATAACTCCAGCTTAGTCTTC CAAAATTCACTTTCATGATGCCACTCAGCATCTCAAATACCTTTCATTGGCTCTCTGCTGCCAAAGGATAAAGGTCAAAGTCATTAGCCT CAACAGTGGGCTTCAACCAGCCTTTGGACCTCAGCCCCATTTATCCATCACAGAGGCTGGTAACTAGTCTCACTGCTCAGGCTGTGAGTG TTCCTGATCTTGTGACATTCTGTGCTGTGCTTTTACATGGAACAGCTCTTTCCTCTCTCTTGGCCCATTCAGATCCTCCTCATCCAGCCC CCATCTGAATCCTGTAACAGACACAACCACATCTACACAGTTTCCACATACCACTTGGAATTGCTGGTTATGCTGCAATGAGAGAATGTC TTTGTCTTTAAATTGTAAGCTTCATGAGGGCAGGGACCATCTGTTTCTCTGTCTCTCCTCACAGTGCCTTGAGCAGTACGTTGTCTGGAA TGTCAGAACCAATAAATACTTGTGGATTGAAACAAAAAGAGAA >12223_12223_2_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000409788_RHBDD1_chr2_227860147_ENST00000341329_length(amino acids)=176AA_BP=1 MVRDETRRVVGYFWYFLFCFFIRNVLLFQNSHALLQILHNVCWRFVRLPKRERHDGSGRGLRCTPPVLPLLQSSSPKSHQAFKTLLSIPR PGSLPGFCEAEVIGSVFPGAVLALRRSFTGLITSFPVSQLFVFSSYLSHQSLSSSGEGTGARFTFESLRCGPTTAPARSLRLGWFH -------------------------------------------------------------- >12223_12223_3_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000410084_RHBDD1_chr2_227860147_ENST00000341329_length(transcript)=4197nt_BP=412nt AGTCGGGCGGGCGGGGAAGTGCGGGCCGGGTCAACTGTCACCGGCTGCACCCGGGCTTCAGCGCCTCGCTCCCGCCCCTGGTAAACTTCC TGTTGTCGGGAGGCAGATACCAGCTTTGTGACCAATTCAGGAAAATGCCTAACTGATTTCTGGTTAGGAATTGGGCGGTCGGAGCGTTCT GGAGTTCAATGGAACCACCCCAAGCGCAGACTGCGTGCAGGCGCCGTAGTCGGCCCGCACCGGAGGGATTCAAAGGTAGCACAGGCGGAG TGCAGCGGAGGCCCCTGCCGCTGCCGTCATGCCGTTCCCGTTTGGGAAGTCTCACAAATCTCCAGCAGACATTGTGAAGAATCTGAAGGA GAGCATGGCTGTTCTGGAAAAGCAAGACATTTCTGATAAAAAAGCAGAAAAGGAAATACCAGAAATAGCCCACCACCCTACGGGTTTCAT CTCTCACCAGAAGAAATGAGGAGACAGCGGCTTCACAGATTCGATAGCCAGTGAGGTGGCATCTTGGGAAGACATGGCCTATTCGTGTAA TTATTGCCCATTTGGCTCATTCCCCAAGCCCCTAATTCATTTTAATTCATTTTAAACAAAAGCAGAGTACACCGGTATTGCTCCAGATCG CTCACATCACCTGGGACAGTCCCATGGCCCCTATGAGTCAACTCACAGCTTGCGGGGAGTGGGCCTTCTCCTGGCCTTGTTCTTGCTCAT AAACAGGTCACTTCCTCCATGAAGAGACCAGTTTCCACGCTCCCATCTCTCACTGCTGACTCAGCGATGCCTCTGCCTCGGTCTGCTTTT GAAGACTGTGACCTTCACCAGGAGGTTTTACTTACACCAGTCGGGAAGATTAGTCCCTCATTCTGCCTGGAGTGCCCCGTGTTTGACTTG GCAGCGGGTGTGGAGCCATCCCCGCGTCCTCCTGGCGCATTGCCACTGTGGCTGTCCAGGAACAGGATGTGGCTGCCTTGGCCGAATGTT GTCCTACTCTCCCCAACCCCGGCGCCTCAGCTCCTCAGCTCCTCGGGCCCCTGCGTCTGGCTGGTGTTTGCAGGGCTTTCGCTCTGCTCT GGTATTGCTCTGCCTTTATAGAAAGTCTTATTGAAGAAGTGTAAGAAAGACCTAAGGTGGGGAAGACTCCTACACACACCATTAGTATCA GTGACACCAGCAATGTAGGTTCCCAGCCCCTTCCCAGTGGCAGCTTGTGTGTCCAGGAGATAGGACATCATTTAACGCATCAGCAAAGTA GCAGCAGATGCCACATACAGAGTAGAGCGAAGGCATTTGGTGGATCGGTCACTAGAGATCTATCTTGCAGAAAGTATGTTTTTCCTCATA AAAGTGCCTCTTAATTGGCCATTGTACCAGCCACTTGTCCTAGCCAAATGTCCAAAACACGCCCTTGGGCCCCGCCACGTTACAATCCAC AGATTGTCTGTCTGAGTCGTTTAAGGCATTTCCTGGTGCTTGTGTTCCATGAATAAAAGGACAAAGTCAGAAGATCACTGATGTCTTACT GTCAACAGAGATATTTTAAAAGAGAGAAGCAGGAAAAGATCTTCCTTTTTTGATCTACAACTTATATAGTTTTCTGATTATGCACATAAT AGATATGCCTTCCAGATGCATAAGGCAAACATCTGGAAAGAAATATACCCAAATCTTAGCAGGGGTTATCTTTGGGAGTGGAGTACATGG GATTTTGCTTTCTTCATTTTTATAATTTTATATTACTGTCTTGGAAGATGTGTTTATGTGTGTGTGTTACTTTTACAATCAGGAAAACAT ATTTAATAACATATAGTCAAGAAAACAGACTTAAAAATAAATACTATGTGTCCATTGAGAAAATTCACAATATAAACAGAAATACAAATA AATACATACACAATTTTAAAGTCACCTGTAGCCCTACCCTTAGAGGTACCCAGGGTTAACATTTTGGTGGTATTGTCTTATCAATTTTTC CGTTGATACATTCAGCAAATTTGGAGCACATTGACCATGGAGTTTTGTGTCCAAATCCAATCTGAATTTACCTGGAAGAGGCCTTGACAC CTGCATGGAAATGAGCTAAGAAAACCACTGGAGCCTTGGGAGCTCTTTGGCCTCCTGGCTGGCCCAGTAATATCTGAGCTCCTTTGGTTA ATTTATAACTGATATAAAACTACATCTTCTTTATAATATAAATTGTACCTGTGAGTCTAGAAGCTTTAAATGTGTTTAAATTAAAATATT CAAGCTAAATGTTACTGCTCTCTCCCAAATTCTGTAAGTTTGACTCCCGTTACCCCAATTAGAAGTAACTTCTTTGTTTCATGCCACTTT TATAGCATTTGGTAATTCTGCTATAACACATCTTGCCCCTATTATTAACTGTGCACAGCTACACAAAGGTGTGCCTTCTACGTGGGAACA TGGATTGTGAATGACTCTGTAATGAGGCCTGAGTCTTAGTTATCTTTCCACTCACTCCCCGTCTCCCCTTTCCAACCCCAAAGGCTCACG ATAGGGGCTCACTAAATGTCAGTGTTTCACCAAAGTATTTTTTCCATTGTATTAAGAGTCCAGTCACTGTATATGGAAGTATTTTATTTT TTATTTTTTTATATCACTTGAGTCCACTAGTAGTACTTCCTTGCTCTGTTTGACTTGTCAGATACAAAGACACGGGATTAGATTTTGGGT GGTAAAATTGTGATACGCATGGCTGTTGATGGAGTGGAACATCTTAGTGATGTGAGAAAGGTCATTTTAGTTATAAATGTAAACCAATTA CTTTAGCACAACAATAAAGATGTTCTGGAAATTATTATAATTATTTTAGTTAATGGACTTGCCTGTAACAAGTGCTTATGTTTGGAATGT CCATTTTATTTTATTTTCCTCTACAATAAGGAATCCCTAAAGTTATGCCAACAAACTGGAATTGGATTGAGAAAGACAAAAGCAAGTGAG CAAGTGTATTAGACTAATTATCTCTAGTAGGGAATAAGCACTTAGAGCTAATTTTTTAATTTGGTTTATTTATTAGTAGCTCCCTGATTG TCTGTTAAGCCCCTGGTTCCCACAGTTTGGTGTGGTCAGTCAACTTAAGCGACCTTTTAATGCAGAATCTTTACTTGGTTCTGAAGAGGA GGCAGTAGTTAAAATTGCTTCCTCTTAATGACTCTGAATGTATTGCCTGTGATTCCAAAGTTGTACATAATTGTTCAGACCTCCTCCATC CTTTTAAATTGCCTGCTGCAGTAAATAACTAGTTTGAGTAGAACTAGATCCTGTCTATCTATTTGGCACATGTTCTGCTGCCTGGGGAGT AAGCAAGCTAAAGGGATGAGAAAGACCACCTCCCCCTACCCTGGAAATTGCACTGCAAGGCAGGGCGAGAATGGGGTAGCTGGCAGACCT GGCCTCCTTGTTCCCAGTCTTAGTTTTTCTTGAGAGATTCAGTATTCAGTAAAGAATAGCATTCAATTAGTCAAAAAATATATATATAAC TTCTTCCTTTCCCTTCCCATGAATCATTGCAGTCATTCCCTAAGTTTCTTCTCTTTTTTTTTCATGGCTGCTAGTATTTTATTTTAGAAT CACAGACTCTCAGCTTAGAGAGTCACAGTATCTCAATTATTTGCTCTGTTCAATACTTAACAATACCTCCTCACTCAATTCTACATAACT CCAGCTTAGTCTTCCAAAATTCACTTTCATGATGCCACTCAGCATCTCAAATACCTTTCATTGGCTCTCTGCTGCCAAAGGATAAAGGTC AAAGTCATTAGCCTCAACAGTGGGCTTCAACCAGCCTTTGGACCTCAGCCCCATTTATCCATCACAGAGGCTGGTAACTAGTCTCACTGC TCAGGCTGTGAGTGTTCCTGATCTTGTGACATTCTGTGCTGTGCTTTTACATGGAACAGCTCTTTCCTCTCTCTTGGCCCATTCAGATCC TCCTCATCCAGCCCCCATCTGAATCCTGTAACAGACACAACCACATCTACACAGTTTCCACATACCACTTGGAATTGCTGGTTATGCTGC AATGAGAGAATGTCTTTGTCTTTAAATTGTAAGCTTCATGAGGGCAGGGACCATCTGTTTCTCTGTCTCTCCTCACAGTGCCTTGAGCAG TACGTTGTCTGGAATGTCAGAACCAATAAATACTTGTGGATTGAAACAAAAAGAGAA >12223_12223_3_CAB39-RHBDD1_CAB39_chr2_231624830_ENST00000410084_RHBDD1_chr2_227860147_ENST00000341329_length(amino acids)=120AA_BP= MKRPVSTLPSLTADSAMPLPRSAFEDCDLHQEVLLTPVGKISPSFCLECPVFDLAAGVEPSPRPPGALPLWLSRNRMWLPWPNVVLLSPT PAPQLLSSSGPCVWLVFAGLSLCSGIALPL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CAB39-RHBDD1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CAB39-RHBDD1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CAB39-RHBDD1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
