
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BRWD1-GNAI3 (FusionGDB2 ID:HG54014TG2773) |
Fusion Gene Summary for BRWD1-GNAI3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BRWD1-GNAI3 | Fusion gene ID: hg54014tg2773 | Hgene | Tgene | Gene symbol | BRWD1 | GNAI3 | Gene ID | 54014 | 2773 |
| Gene name | bromodomain and WD repeat domain containing 1 | G protein subunit alpha i3 | |
| Synonyms | C21orf107|DCAF19|N143|WDR9|WRD9 | 87U6|ARCND1 | |
| Cytomap | ('BRWD1')('GNAI3') 21q22.2 | 1p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | bromodomain and WD repeat-containing protein 1WD repeat protein WDR9-form2WD repeat-containing protein 9transcriptional unit N143 | guanine nucleotide-binding protein G(i) subunit alphag(i) alpha-3guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3guanine nucleotide-binding protein G(k) subunit alpha | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | . | P08754 | |
| Ensembl transtripts involved in fusion gene | ENST00000333229, ENST00000342449, ENST00000380800, ENST00000341322, ENST00000470108, | ||
| Fusion gene scores | * DoF score | 28 X 28 X 10=7840 | 33 X 8 X 14=3696 |
| # samples | 32 | 35 | |
| ** MAII score | log2(32/7840*10)=-4.61470984411521 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(35/3696*10)=-3.40053792958373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: BRWD1 [Title/Abstract] AND GNAI3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BRWD1(40670357)-GNAI3(110116358), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | BRWD1-GNAI3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BRWD1-GNAI3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BRWD1-GNAI3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BRWD1-GNAI3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GNAI3 | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 19478087 |
| Tgene | GNAI3 | GO:0007212 | dopamine receptor signaling pathway | 19478087 |
| Tgene | GNAI3 | GO:0046039 | GTP metabolic process | 19478087 |
| Tgene | GNAI3 | GO:0051301 | cell division | 17635935 |
 Fusion gene breakpoints across BRWD1 (5'-gene) Fusion gene breakpoints across BRWD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
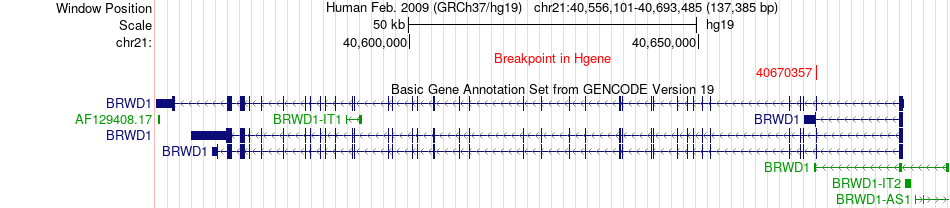 |
 Fusion gene breakpoints across GNAI3 (3'-gene) Fusion gene breakpoints across GNAI3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
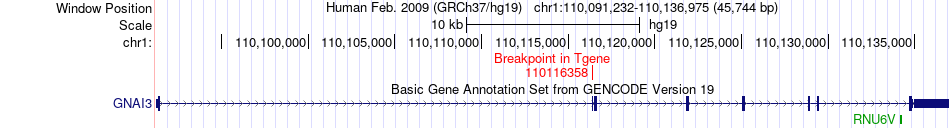 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A1XT | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
Top |
Fusion Gene ORF analysis for BRWD1-GNAI3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000333229 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
| In-frame | ENST00000342449 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
| In-frame | ENST00000380800 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
| intron-3CDS | ENST00000341322 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
| intron-3CDS | ENST00000470108 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000333229 | BRWD1 | chr21 | 40670357 | - | ENST00000369851 | GNAI3 | chr1 | 110116358 | + | 3655 | 677 | 235 | 1623 | 462 |
| ENST00000342449 | BRWD1 | chr21 | 40670357 | - | ENST00000369851 | GNAI3 | chr1 | 110116358 | + | 3406 | 428 | 79 | 1374 | 431 |
| ENST00000380800 | BRWD1 | chr21 | 40670357 | - | ENST00000369851 | GNAI3 | chr1 | 110116358 | + | 3426 | 448 | 6 | 1394 | 462 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000333229 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + | 0.000203461 | 0.99979657 |
| ENST00000342449 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + | 0.000186399 | 0.9998136 |
| ENST00000380800 | ENST00000369851 | BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + | 0.000184975 | 0.99981505 |
Top |
Fusion Genomic Features for BRWD1-GNAI3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + | 3.14E-09 | 1 |
| BRWD1 | chr21 | 40670357 | - | GNAI3 | chr1 | 110116358 | + | 3.14E-09 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
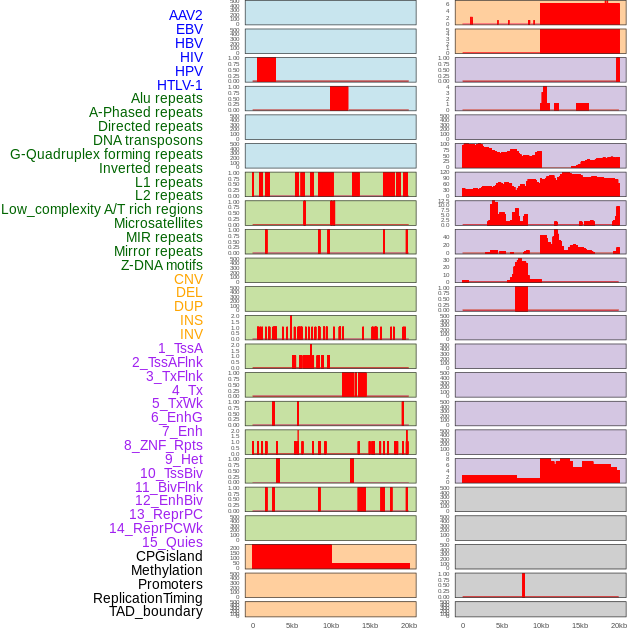 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
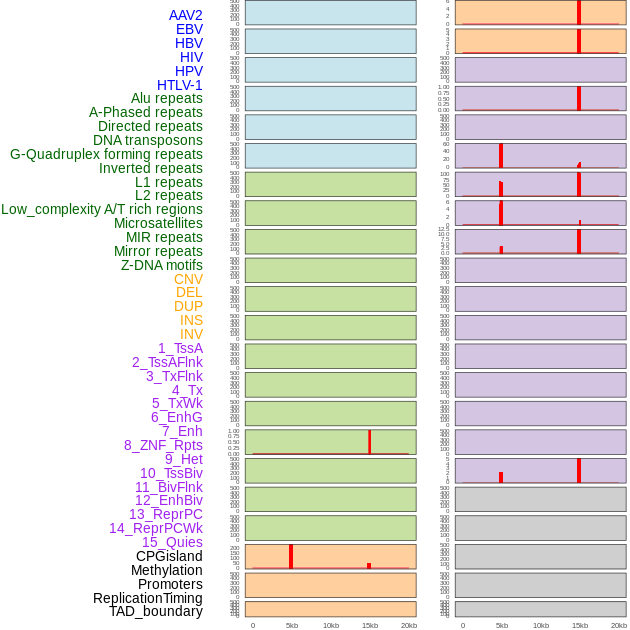 |
Top |
Fusion Protein Features for BRWD1-GNAI3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:40670357/chr1:110116358) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | GNAI3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Heterotrimeric guanine nucleotide-binding proteins (G proteins) function as transducers downstream of G protein-coupled receptors (GPCRs) in numerous signaling cascades. The alpha chain contains the guanine nucleotide binding site and alternates between an active, GTP-bound state and an inactive, GDP-bound state. Signaling by an activated GPCR promotes GDP release and GTP binding. The alpha subunit has a low GTPase activity that converts bound GTP to GDP, thereby terminating the signal. Both GDP release and GTP hydrolysis are modulated by numerous regulatory proteins (PubMed:8774883, PubMed:18434541, PubMed:19478087). Signaling is mediated via effector proteins, such as adenylate cyclase. Inhibits adenylate cyclase activity, leading to decreased intracellular cAMP levels (PubMed:19478087). Stimulates the activity of receptor-regulated K(+) channels (PubMed:2535845). The active GTP-bound form prevents the association of RGS14 with centrosomes and is required for the translocation of RGS14 from the cytoplasm to the plasma membrane. May play a role in cell division (PubMed:17635935). {ECO:0000269|PubMed:17635935, ECO:0000269|PubMed:18434541, ECO:0000269|PubMed:2535845, ECO:0000269|PubMed:8774883}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 150_151 | 39 | 948.6666666666666 | Nucleotide binding | GTP | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 175_181 | 39 | 948.6666666666666 | Nucleotide binding | GTP | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 200_204 | 39 | 948.6666666666666 | Nucleotide binding | GTP | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 269_272 | 39 | 948.6666666666666 | Nucleotide binding | GTP | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 43_48 | 39 | 948.6666666666666 | Nucleotide binding | GTP | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 173_181 | 39 | 948.6666666666666 | Region | G2 motif | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 196_205 | 39 | 948.6666666666666 | Region | G3 motif | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 265_272 | 39 | 948.6666666666666 | Region | G4 motif | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 324_329 | 39 | 948.6666666666666 | Region | G5 motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 1539_1548 | 116 | 2321.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 1687_1692 | 116 | 2321.0 | Compositional bias | Note=Poly-Glu |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 661_668 | 116 | 2321.0 | Compositional bias | Note=Poly-Gln |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 821_824 | 116 | 2321.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 1539_1548 | 0 | 121.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 1687_1692 | 0 | 121.0 | Compositional bias | Note=Poly-Glu |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 661_668 | 0 | 121.0 | Compositional bias | Note=Poly-Gln |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 821_824 | 0 | 121.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 1539_1548 | 116 | 2270.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 1687_1692 | 116 | 2270.0 | Compositional bias | Note=Poly-Glu |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 661_668 | 116 | 2270.0 | Compositional bias | Note=Poly-Gln |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 821_824 | 116 | 2270.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 1539_1548 | 116 | 2200.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 1687_1692 | 116 | 2200.0 | Compositional bias | Note=Poly-Glu |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 661_668 | 116 | 2200.0 | Compositional bias | Note=Poly-Gln |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 821_824 | 116 | 2200.0 | Compositional bias | Note=Poly-Ser |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 1177_1247 | 116 | 2321.0 | Domain | Bromo 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 1330_1400 | 116 | 2321.0 | Domain | Bromo 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 1177_1247 | 0 | 121.0 | Domain | Bromo 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 1330_1400 | 0 | 121.0 | Domain | Bromo 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 1177_1247 | 116 | 2270.0 | Domain | Bromo 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 1330_1400 | 116 | 2270.0 | Domain | Bromo 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 1177_1247 | 116 | 2200.0 | Domain | Bromo 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 1330_1400 | 116 | 2200.0 | Domain | Bromo 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 184_223 | 116 | 2321.0 | Repeat | Note=WD 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 226_265 | 116 | 2321.0 | Repeat | Note=WD 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 268_311 | 116 | 2321.0 | Repeat | Note=WD 3 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 322_365 | 116 | 2321.0 | Repeat | Note=WD 4 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 366_405 | 116 | 2321.0 | Repeat | Note=WD 5 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 424_463 | 116 | 2321.0 | Repeat | Note=WD 6 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 466_506 | 116 | 2321.0 | Repeat | Note=WD 7 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000333229 | - | 5 | 42 | 514_553 | 116 | 2321.0 | Repeat | Note=WD 8 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 184_223 | 0 | 121.0 | Repeat | Note=WD 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 226_265 | 0 | 121.0 | Repeat | Note=WD 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 268_311 | 0 | 121.0 | Repeat | Note=WD 3 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 322_365 | 0 | 121.0 | Repeat | Note=WD 4 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 366_405 | 0 | 121.0 | Repeat | Note=WD 5 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 424_463 | 0 | 121.0 | Repeat | Note=WD 6 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 466_506 | 0 | 121.0 | Repeat | Note=WD 7 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000341322 | - | 1 | 5 | 514_553 | 0 | 121.0 | Repeat | Note=WD 8 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 184_223 | 116 | 2270.0 | Repeat | Note=WD 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 226_265 | 116 | 2270.0 | Repeat | Note=WD 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 268_311 | 116 | 2270.0 | Repeat | Note=WD 3 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 322_365 | 116 | 2270.0 | Repeat | Note=WD 4 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 366_405 | 116 | 2270.0 | Repeat | Note=WD 5 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 424_463 | 116 | 2270.0 | Repeat | Note=WD 6 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 466_506 | 116 | 2270.0 | Repeat | Note=WD 7 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000342449 | - | 5 | 41 | 514_553 | 116 | 2270.0 | Repeat | Note=WD 8 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 184_223 | 116 | 2200.0 | Repeat | Note=WD 1 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 226_265 | 116 | 2200.0 | Repeat | Note=WD 2 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 268_311 | 116 | 2200.0 | Repeat | Note=WD 3 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 322_365 | 116 | 2200.0 | Repeat | Note=WD 4 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 366_405 | 116 | 2200.0 | Repeat | Note=WD 5 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 424_463 | 116 | 2200.0 | Repeat | Note=WD 6 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 466_506 | 116 | 2200.0 | Repeat | Note=WD 7 |
| Hgene | BRWD1 | chr21:40670357 | chr1:110116358 | ENST00000380800 | - | 5 | 42 | 514_553 | 116 | 2200.0 | Repeat | Note=WD 8 |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 32_354 | 39 | 948.6666666666666 | Domain | G-alpha | |
| Tgene | GNAI3 | chr21:40670357 | chr1:110116358 | ENST00000369851 | 0 | 9 | 35_48 | 39 | 948.6666666666666 | Region | G1 motif |
Top |
Fusion Gene Sequence for BRWD1-GNAI3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >10346_10346_1_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000333229_GNAI3_chr1_110116358_ENST00000369851_length(transcript)=3655nt_BP=677nt AGGTCTTTGCCTCCGCGTCACTTTCTCCTCATCGAGTCCCCGGAGGCAGAGAGCTGAGCGAGGTTCGCCTCCCCGGGCGGGACACGAGCT CCGGTCCTCTCTTGGCGCAAACCCCCGCCCCCCGCGGAGGGACTACTTTTCGTCTCCCGCGGAGGAACTACTTCCTCCCCCGCGGAGGCG TCGGTCCAGGCGGCGCGCACGGTATGGCGGCGGCGGCGGCGGCGGCGGCGCGCGCCTGAGAAGAGGGGACGCCAGCCCGGTCAGGCCTCG GCGCGGCCTACACGCTCCGCTCGCTCGCTCGCTCCCGCCTCCCGCCCCGCGCCCGGCCATGGCGGAGCCGTCGTCCGCCCGACGCCCGGT GCCTCTCATCGAGTCGGAGCTGTACTTCCTTATCGCCCGGTACCTATCGGCGGGCCCGTGTCGGAGAGCGGCCCAGGTGCTGGTGCAGGA GCTGGAGCAGTACCAGTTGTTGCCGAAGAGATTGGACTGGGAGGGCAACGAGCACAACAGGAGCTACGAGGAGTTGGTCTTGTCCAATAA GCATGTGGCTCCTGATCATCTTTTGCAAATCTGCCAGCGCATCGGTCCTATGTTGGATAAAGAAATTCCACCCAGTATTTCAAGAGTCAC TTCTTTACTTGGTGCAGGAAGGCAGTCTTTGCTACGTACAGCAAAAGGTGCTGGAGAATCTGGTAAAAGCACCATTGTGAAACAGATGAA AATCATTCATGAGGATGGCTATTCAGAGGATGAATGTAAACAATATAAAGTAGTTGTCTACAGCAATACTATACAGTCCATCATTGCAAT CATAAGAGCCATGGGACGGCTAAAGATTGACTTTGGGGAAGCTGCCAGGGCAGATGATGCCCGGCAATTATTTGTTTTAGCTGGCAGTGC TGAAGAAGGAGTCATGACTCCAGAACTAGCAGGAGTGATTAAACGGTTATGGCGAGATGGTGGGGTACAAGCTTGCTTCAGCAGATCCAG GGAATATCAGCTCAATGATTCTGCTTCATATTATCTAAATGATCTGGATAGAATATCCCAGTCTAACTACATTCCAACTCAGCAAGATGT TCTTCGGACGAGAGTGAAGACCACAGGCATTGTAGAAACACATTTCACCTTCAAAGACCTATACTTCAAGATGTTTGATGTAGGTGGCCA AAGATCAGAACGAAAAAAGTGGATTCACTGTTTTGAGGGAGTGACAGCAATTATCTTCTGTGTGGCCCTCAGTGATTATGACCTTGTTCT GGCTGAGGACGAGGAGATGAACCGAATGCATGAAAGCATGAAACTGTTTGACAGCATTTGTAATAACAAATGGTTTACAGAAACTTCAAT CATTCTCTTCCTTAACAAGAAAGACCTTTTTGAGGAAAAAATAAAGAGGAGTCCGTTAACTATCTGTTATCCAGAATACACAGGTTCCAA TACATATGAAGAGGCAGCTGCCTATATTCAATGCCAGTTTGAAGATCTGAACAGAAGAAAAGATACCAAGGAGATCTATACTCACTTCAC CTGTGCCACAGACACGAAGAATGTGCAGTTTGTTTTTGATGCTGTTACAGATGTCATCATTAAAAACAACTTAAAGGAATGTGGACTTTA TTGAGAAGCATGGATGTTAGTGAAAGTTACTACAGTGTGGAGTGTTGAGACCAGACACCTTTTGCTGTCTCATGGGGCAGCTACAAGCAT GAACGGGACCAGGGAATGGCAGCAGCATGCAGAATCTTAGCACTCTTTAGCACAATATTTTGTATTAGGGAACTTTTAATTGACATGAGA TGCTAAAGTCAGACATTGGAATTGGAAGAACTATAAAGTGTGATTCGATCGTCAAGACATCACTTGGATTTCTTAATCTTAAATGCTTAT GGAAGATGTGAAGTTGAGGTGCTGCATTCTAGAACTTCAATATGTAGCTTACTCTTTTTTTCCCCCCTTCTTAAACCACCAGTGGTTCAT TTTTAAGGTTTTTTCATCAAGAGAAGAATAACTTTACTAAATTTTATTTCTTTATTTGCAAAAGAATCTTTATTAAAACAAACAATCTTA ACTATGCACATGATGTGACCAGATCATCTTGAAAATATTCCTCTTTAGTAGGAACTCTTTGTTTTTAACTCTTGGTATGGTCAGAATATA ATACTTCCATAATTACTTATAATTCTTTCCGGTTACTGGGGCTATAAATACAACTTTTTAAATGAAATTTATGTTATTATCCTGCTCAAA GTACCATTATGGTTTCCATATGGTAATTACATTGGAAAGTTCTGCAATAAGATTCTAGTTCTCTCTTTTCTTTAAGCTTATGGTTGAAGG TTAACCTTGTTTGTGCAGATTACCAAATTACTGCTGATGTAGTATGATAAAGTCAGCTTCTTGGATACAGACACACCCAGGGCAGCTGCC AATCAGAAATGCAAATTGCTAAATTCCAAACAGAACCAGTGACTTTGCTGCTACATATTTACTAAATTGACCACTTTGATTCTTGCTTGT TTGTCTCAGTTATAGGGAGTTTTGTACATGGTGCCTCTTGTTTTCCATCTTCATGGCCTTTTCTGTTGAGAAACATCTAAAGTGTATTAA CAGTGCATGCTTTTACTTTGTAAATGTGGTGCCAAATCCCTGTTTGCCATCGTTTTTACTGTACCGATGTTAACTGTAGTAATCCTTAGC CAGTATGTTCTTTTGCTGAAACTGTTGCATTTTGGGACTTTTTTCCACTTTGTCATGTGTACATTTTTAATCTGTTATAGTTGGCGGCAG TATTAAAATGGTGAGTAAAGAGCCCAGGTTTGCTCCTGTTTGTAACTAGTCTTTTCTGGAAATACTGTCTTCTGTTTCCTTTTGCCTTTG CAGAATGTGCTTTAGCCTTTGTCTGAGAATGGGTTTAGGTAGAAGGGTTTCCTGAAAGTCTAGTCTTTTGGTGTAATAGTATTGTTGAAC CTTAGGTTTTATGTTATATCTGCATATGAGTGATATGTGATCATGATTCATTTTGCAGATTTACATTTTGCTTGTGTGGAAAGTTACGTT CACTTCAACCTACAGACCCTTTTGTATAATGTACAGCAAATGTCATTAAATATTGAATGCTCTATTGGGGGAAGACAAATGAAGAGAATG CATTTTGAGCATTTCAAATCAGTAGTTTGGGCAGTGCCTTTTGGGGTCCAATCTTTTTGGATTGGATTCTATTTCATCTTTTGATGTGAC TTTTCACTAGTTTACAAAAAAAAGGTTGGTGGTCAACAAAAAAAGCAAGTCATAGCAGTTCAGTGCCTTTGTTGGTAGTTTTTAACTTTT TTTGTGATTCTTTTTGGAAAATCATAGCTTACAGATGGTATAACTGTATTATATAATGGAATTTTCTTTAGGTGTGGGAAAACTTGATCT GAAAGAAAATTATCAGCTTCAATTGGCGATTGATTCAGTGCCCACAATGTAAACAGGGTTGGTAGTTGTTACTCATTTTGAATATACCTT TTCCTTATTGTATTCTGTAATATAGGATCCTGGAAATGAGACCTGGTGGAATATTATTTTCAACTGTAGTCTTTCTATACTGTGGACTAA TGAAATGGTGTTGTTGATAATCTAAAATAACCAATAATCCAAAAAAGTTCATTTT >10346_10346_1_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000333229_GNAI3_chr1_110116358_ENST00000369851_length(amino acids)=462AA_BP=147 MRRGDASPVRPRRGLHAPLARSLPPPAPRPAMAEPSSARRPVPLIESELYFLIARYLSAGPCRRAAQVLVQELEQYQLLPKRLDWEGNEH NRSYEELVLSNKHVAPDHLLQICQRIGPMLDKEIPPSISRVTSLLGAGRQSLLRTAKGAGESGKSTIVKQMKIIHEDGYSEDECKQYKVV VYSNTIQSIIAIIRAMGRLKIDFGEAARADDARQLFVLAGSAEEGVMTPELAGVIKRLWRDGGVQACFSRSREYQLNDSASYYLNDLDRI SQSNYIPTQQDVLRTRVKTTGIVETHFTFKDLYFKMFDVGGQRSERKKWIHCFEGVTAIIFCVALSDYDLVLAEDEEMNRMHESMKLFDS ICNNKWFTETSIILFLNKKDLFEEKIKRSPLTICYPEYTGSNTYEEAAAYIQCQFEDLNRRKDTKEIYTHFTCATDTKNVQFVFDAVTDV IIKNNLKECGLY -------------------------------------------------------------- >10346_10346_2_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000342449_GNAI3_chr1_110116358_ENST00000369851_length(transcript)=3406nt_BP=428nt CGCCAGCCCGGTCAGGCCTCGGCGCGGCCTACACGCTCCGCTCGCTCGCTCGCTCCCGCCTCCCGCCCCGCGCCCGGCCATGGCGGAGCC GTCGTCCGCCCGACGCCCGGTGCCTCTCATCGAGTCGGAGCTGTACTTCCTTATCGCCCGGTACCTATCGGCGGGCCCGTGTCGGAGAGC GGCCCAGGTGCTGGTGCAGGAGCTGGAGCAGTACCAGTTGTTGCCGAAGAGATTGGACTGGGAGGGCAACGAGCACAACAGGAGCTACGA GGAGTTGGTCTTGTCCAATAAGCATGTGGCTCCTGATCATCTTTTGCAAATCTGCCAGCGCATCGGTCCTATGTTGGATAAAGAAATTCC ACCCAGTATTTCAAGAGTCACTTCTTTACTTGGTGCAGGAAGGCAGTCTTTGCTACGTACAGCAAAAGGTGCTGGAGAATCTGGTAAAAG CACCATTGTGAAACAGATGAAAATCATTCATGAGGATGGCTATTCAGAGGATGAATGTAAACAATATAAAGTAGTTGTCTACAGCAATAC TATACAGTCCATCATTGCAATCATAAGAGCCATGGGACGGCTAAAGATTGACTTTGGGGAAGCTGCCAGGGCAGATGATGCCCGGCAATT ATTTGTTTTAGCTGGCAGTGCTGAAGAAGGAGTCATGACTCCAGAACTAGCAGGAGTGATTAAACGGTTATGGCGAGATGGTGGGGTACA AGCTTGCTTCAGCAGATCCAGGGAATATCAGCTCAATGATTCTGCTTCATATTATCTAAATGATCTGGATAGAATATCCCAGTCTAACTA CATTCCAACTCAGCAAGATGTTCTTCGGACGAGAGTGAAGACCACAGGCATTGTAGAAACACATTTCACCTTCAAAGACCTATACTTCAA GATGTTTGATGTAGGTGGCCAAAGATCAGAACGAAAAAAGTGGATTCACTGTTTTGAGGGAGTGACAGCAATTATCTTCTGTGTGGCCCT CAGTGATTATGACCTTGTTCTGGCTGAGGACGAGGAGATGAACCGAATGCATGAAAGCATGAAACTGTTTGACAGCATTTGTAATAACAA ATGGTTTACAGAAACTTCAATCATTCTCTTCCTTAACAAGAAAGACCTTTTTGAGGAAAAAATAAAGAGGAGTCCGTTAACTATCTGTTA TCCAGAATACACAGGTTCCAATACATATGAAGAGGCAGCTGCCTATATTCAATGCCAGTTTGAAGATCTGAACAGAAGAAAAGATACCAA GGAGATCTATACTCACTTCACCTGTGCCACAGACACGAAGAATGTGCAGTTTGTTTTTGATGCTGTTACAGATGTCATCATTAAAAACAA CTTAAAGGAATGTGGACTTTATTGAGAAGCATGGATGTTAGTGAAAGTTACTACAGTGTGGAGTGTTGAGACCAGACACCTTTTGCTGTC TCATGGGGCAGCTACAAGCATGAACGGGACCAGGGAATGGCAGCAGCATGCAGAATCTTAGCACTCTTTAGCACAATATTTTGTATTAGG GAACTTTTAATTGACATGAGATGCTAAAGTCAGACATTGGAATTGGAAGAACTATAAAGTGTGATTCGATCGTCAAGACATCACTTGGAT TTCTTAATCTTAAATGCTTATGGAAGATGTGAAGTTGAGGTGCTGCATTCTAGAACTTCAATATGTAGCTTACTCTTTTTTTCCCCCCTT CTTAAACCACCAGTGGTTCATTTTTAAGGTTTTTTCATCAAGAGAAGAATAACTTTACTAAATTTTATTTCTTTATTTGCAAAAGAATCT TTATTAAAACAAACAATCTTAACTATGCACATGATGTGACCAGATCATCTTGAAAATATTCCTCTTTAGTAGGAACTCTTTGTTTTTAAC TCTTGGTATGGTCAGAATATAATACTTCCATAATTACTTATAATTCTTTCCGGTTACTGGGGCTATAAATACAACTTTTTAAATGAAATT TATGTTATTATCCTGCTCAAAGTACCATTATGGTTTCCATATGGTAATTACATTGGAAAGTTCTGCAATAAGATTCTAGTTCTCTCTTTT CTTTAAGCTTATGGTTGAAGGTTAACCTTGTTTGTGCAGATTACCAAATTACTGCTGATGTAGTATGATAAAGTCAGCTTCTTGGATACA GACACACCCAGGGCAGCTGCCAATCAGAAATGCAAATTGCTAAATTCCAAACAGAACCAGTGACTTTGCTGCTACATATTTACTAAATTG ACCACTTTGATTCTTGCTTGTTTGTCTCAGTTATAGGGAGTTTTGTACATGGTGCCTCTTGTTTTCCATCTTCATGGCCTTTTCTGTTGA GAAACATCTAAAGTGTATTAACAGTGCATGCTTTTACTTTGTAAATGTGGTGCCAAATCCCTGTTTGCCATCGTTTTTACTGTACCGATG TTAACTGTAGTAATCCTTAGCCAGTATGTTCTTTTGCTGAAACTGTTGCATTTTGGGACTTTTTTCCACTTTGTCATGTGTACATTTTTA ATCTGTTATAGTTGGCGGCAGTATTAAAATGGTGAGTAAAGAGCCCAGGTTTGCTCCTGTTTGTAACTAGTCTTTTCTGGAAATACTGTC TTCTGTTTCCTTTTGCCTTTGCAGAATGTGCTTTAGCCTTTGTCTGAGAATGGGTTTAGGTAGAAGGGTTTCCTGAAAGTCTAGTCTTTT GGTGTAATAGTATTGTTGAACCTTAGGTTTTATGTTATATCTGCATATGAGTGATATGTGATCATGATTCATTTTGCAGATTTACATTTT GCTTGTGTGGAAAGTTACGTTCACTTCAACCTACAGACCCTTTTGTATAATGTACAGCAAATGTCATTAAATATTGAATGCTCTATTGGG GGAAGACAAATGAAGAGAATGCATTTTGAGCATTTCAAATCAGTAGTTTGGGCAGTGCCTTTTGGGGTCCAATCTTTTTGGATTGGATTC TATTTCATCTTTTGATGTGACTTTTCACTAGTTTACAAAAAAAAGGTTGGTGGTCAACAAAAAAAGCAAGTCATAGCAGTTCAGTGCCTT TGTTGGTAGTTTTTAACTTTTTTTGTGATTCTTTTTGGAAAATCATAGCTTACAGATGGTATAACTGTATTATATAATGGAATTTTCTTT AGGTGTGGGAAAACTTGATCTGAAAGAAAATTATCAGCTTCAATTGGCGATTGATTCAGTGCCCACAATGTAAACAGGGTTGGTAGTTGT TACTCATTTTGAATATACCTTTTCCTTATTGTATTCTGTAATATAGGATCCTGGAAATGAGACCTGGTGGAATATTATTTTCAACTGTAG TCTTTCTATACTGTGGACTAATGAAATGGTGTTGTTGATAATCTAAAATAACCAATAATCCAAAAAAGTTCATTTT >10346_10346_2_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000342449_GNAI3_chr1_110116358_ENST00000369851_length(amino acids)=431AA_BP=116 MAEPSSARRPVPLIESELYFLIARYLSAGPCRRAAQVLVQELEQYQLLPKRLDWEGNEHNRSYEELVLSNKHVAPDHLLQICQRIGPMLD KEIPPSISRVTSLLGAGRQSLLRTAKGAGESGKSTIVKQMKIIHEDGYSEDECKQYKVVVYSNTIQSIIAIIRAMGRLKIDFGEAARADD ARQLFVLAGSAEEGVMTPELAGVIKRLWRDGGVQACFSRSREYQLNDSASYYLNDLDRISQSNYIPTQQDVLRTRVKTTGIVETHFTFKD LYFKMFDVGGQRSERKKWIHCFEGVTAIIFCVALSDYDLVLAEDEEMNRMHESMKLFDSICNNKWFTETSIILFLNKKDLFEEKIKRSPL TICYPEYTGSNTYEEAAAYIQCQFEDLNRRKDTKEIYTHFTCATDTKNVQFVFDAVTDVIIKNNLKECGLY -------------------------------------------------------------- >10346_10346_3_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000380800_GNAI3_chr1_110116358_ENST00000369851_length(transcript)=3426nt_BP=448nt GCGCGCCTGAGAAGAGGGGACGCCAGCCCGGTCAGGCCTCGGCGCGGCCTACACGCTCCGCTCGCTCGCTCGCTCCCGCCTCCCGCCCCG CGCCCGGCCATGGCGGAGCCGTCGTCCGCCCGACGCCCGGTGCCTCTCATCGAGTCGGAGCTGTACTTCCTTATCGCCCGGTACCTATCG GCGGGCCCGTGTCGGAGAGCGGCCCAGGTGCTGGTGCAGGAGCTGGAGCAGTACCAGTTGTTGCCGAAGAGATTGGACTGGGAGGGCAAC GAGCACAACAGGAGCTACGAGGAGTTGGTCTTGTCCAATAAGCATGTGGCTCCTGATCATCTTTTGCAAATCTGCCAGCGCATCGGTCCT ATGTTGGATAAAGAAATTCCACCCAGTATTTCAAGAGTCACTTCTTTACTTGGTGCAGGAAGGCAGTCTTTGCTACGTACAGCAAAAGGT GCTGGAGAATCTGGTAAAAGCACCATTGTGAAACAGATGAAAATCATTCATGAGGATGGCTATTCAGAGGATGAATGTAAACAATATAAA GTAGTTGTCTACAGCAATACTATACAGTCCATCATTGCAATCATAAGAGCCATGGGACGGCTAAAGATTGACTTTGGGGAAGCTGCCAGG GCAGATGATGCCCGGCAATTATTTGTTTTAGCTGGCAGTGCTGAAGAAGGAGTCATGACTCCAGAACTAGCAGGAGTGATTAAACGGTTA TGGCGAGATGGTGGGGTACAAGCTTGCTTCAGCAGATCCAGGGAATATCAGCTCAATGATTCTGCTTCATATTATCTAAATGATCTGGAT AGAATATCCCAGTCTAACTACATTCCAACTCAGCAAGATGTTCTTCGGACGAGAGTGAAGACCACAGGCATTGTAGAAACACATTTCACC TTCAAAGACCTATACTTCAAGATGTTTGATGTAGGTGGCCAAAGATCAGAACGAAAAAAGTGGATTCACTGTTTTGAGGGAGTGACAGCA ATTATCTTCTGTGTGGCCCTCAGTGATTATGACCTTGTTCTGGCTGAGGACGAGGAGATGAACCGAATGCATGAAAGCATGAAACTGTTT GACAGCATTTGTAATAACAAATGGTTTACAGAAACTTCAATCATTCTCTTCCTTAACAAGAAAGACCTTTTTGAGGAAAAAATAAAGAGG AGTCCGTTAACTATCTGTTATCCAGAATACACAGGTTCCAATACATATGAAGAGGCAGCTGCCTATATTCAATGCCAGTTTGAAGATCTG AACAGAAGAAAAGATACCAAGGAGATCTATACTCACTTCACCTGTGCCACAGACACGAAGAATGTGCAGTTTGTTTTTGATGCTGTTACA GATGTCATCATTAAAAACAACTTAAAGGAATGTGGACTTTATTGAGAAGCATGGATGTTAGTGAAAGTTACTACAGTGTGGAGTGTTGAG ACCAGACACCTTTTGCTGTCTCATGGGGCAGCTACAAGCATGAACGGGACCAGGGAATGGCAGCAGCATGCAGAATCTTAGCACTCTTTA GCACAATATTTTGTATTAGGGAACTTTTAATTGACATGAGATGCTAAAGTCAGACATTGGAATTGGAAGAACTATAAAGTGTGATTCGAT CGTCAAGACATCACTTGGATTTCTTAATCTTAAATGCTTATGGAAGATGTGAAGTTGAGGTGCTGCATTCTAGAACTTCAATATGTAGCT TACTCTTTTTTTCCCCCCTTCTTAAACCACCAGTGGTTCATTTTTAAGGTTTTTTCATCAAGAGAAGAATAACTTTACTAAATTTTATTT CTTTATTTGCAAAAGAATCTTTATTAAAACAAACAATCTTAACTATGCACATGATGTGACCAGATCATCTTGAAAATATTCCTCTTTAGT AGGAACTCTTTGTTTTTAACTCTTGGTATGGTCAGAATATAATACTTCCATAATTACTTATAATTCTTTCCGGTTACTGGGGCTATAAAT ACAACTTTTTAAATGAAATTTATGTTATTATCCTGCTCAAAGTACCATTATGGTTTCCATATGGTAATTACATTGGAAAGTTCTGCAATA AGATTCTAGTTCTCTCTTTTCTTTAAGCTTATGGTTGAAGGTTAACCTTGTTTGTGCAGATTACCAAATTACTGCTGATGTAGTATGATA AAGTCAGCTTCTTGGATACAGACACACCCAGGGCAGCTGCCAATCAGAAATGCAAATTGCTAAATTCCAAACAGAACCAGTGACTTTGCT GCTACATATTTACTAAATTGACCACTTTGATTCTTGCTTGTTTGTCTCAGTTATAGGGAGTTTTGTACATGGTGCCTCTTGTTTTCCATC TTCATGGCCTTTTCTGTTGAGAAACATCTAAAGTGTATTAACAGTGCATGCTTTTACTTTGTAAATGTGGTGCCAAATCCCTGTTTGCCA TCGTTTTTACTGTACCGATGTTAACTGTAGTAATCCTTAGCCAGTATGTTCTTTTGCTGAAACTGTTGCATTTTGGGACTTTTTTCCACT TTGTCATGTGTACATTTTTAATCTGTTATAGTTGGCGGCAGTATTAAAATGGTGAGTAAAGAGCCCAGGTTTGCTCCTGTTTGTAACTAG TCTTTTCTGGAAATACTGTCTTCTGTTTCCTTTTGCCTTTGCAGAATGTGCTTTAGCCTTTGTCTGAGAATGGGTTTAGGTAGAAGGGTT TCCTGAAAGTCTAGTCTTTTGGTGTAATAGTATTGTTGAACCTTAGGTTTTATGTTATATCTGCATATGAGTGATATGTGATCATGATTC ATTTTGCAGATTTACATTTTGCTTGTGTGGAAAGTTACGTTCACTTCAACCTACAGACCCTTTTGTATAATGTACAGCAAATGTCATTAA ATATTGAATGCTCTATTGGGGGAAGACAAATGAAGAGAATGCATTTTGAGCATTTCAAATCAGTAGTTTGGGCAGTGCCTTTTGGGGTCC AATCTTTTTGGATTGGATTCTATTTCATCTTTTGATGTGACTTTTCACTAGTTTACAAAAAAAAGGTTGGTGGTCAACAAAAAAAGCAAG TCATAGCAGTTCAGTGCCTTTGTTGGTAGTTTTTAACTTTTTTTGTGATTCTTTTTGGAAAATCATAGCTTACAGATGGTATAACTGTAT TATATAATGGAATTTTCTTTAGGTGTGGGAAAACTTGATCTGAAAGAAAATTATCAGCTTCAATTGGCGATTGATTCAGTGCCCACAATG TAAACAGGGTTGGTAGTTGTTACTCATTTTGAATATACCTTTTCCTTATTGTATTCTGTAATATAGGATCCTGGAAATGAGACCTGGTGG AATATTATTTTCAACTGTAGTCTTTCTATACTGTGGACTAATGAAATGGTGTTGTTGATAATCTAAAATAACCAATAATCCAAAAAAGTT CATTTT >10346_10346_3_BRWD1-GNAI3_BRWD1_chr21_40670357_ENST00000380800_GNAI3_chr1_110116358_ENST00000369851_length(amino acids)=462AA_BP=147 MRRGDASPVRPRRGLHAPLARSLPPPAPRPAMAEPSSARRPVPLIESELYFLIARYLSAGPCRRAAQVLVQELEQYQLLPKRLDWEGNEH NRSYEELVLSNKHVAPDHLLQICQRIGPMLDKEIPPSISRVTSLLGAGRQSLLRTAKGAGESGKSTIVKQMKIIHEDGYSEDECKQYKVV VYSNTIQSIIAIIRAMGRLKIDFGEAARADDARQLFVLAGSAEEGVMTPELAGVIKRLWRDGGVQACFSRSREYQLNDSASYYLNDLDRI SQSNYIPTQQDVLRTRVKTTGIVETHFTFKDLYFKMFDVGGQRSERKKWIHCFEGVTAIIFCVALSDYDLVLAEDEEMNRMHESMKLFDS ICNNKWFTETSIILFLNKKDLFEEKIKRSPLTICYPEYTGSNTYEEAAAYIQCQFEDLNRRKDTKEIYTHFTCATDTKNVQFVFDAVTDV IIKNNLKECGLY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BRWD1-GNAI3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BRWD1-GNAI3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BRWD1-GNAI3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | BRWD1 | C0028960 | Oligospermia | 1 | CTD_human |
| Tgene | C4551996 | Auriculocondylar syndrome 1 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0002622 | Amnesia | 1 | CTD_human | |
| Tgene | C0078917 | Albinism, Ocular | 1 | GENOMICS_ENGLAND | |
| Tgene | C0152423 | Congenital small ears | 1 | GENOMICS_ENGLAND | |
| Tgene | C0233750 | Hysterical amnesia | 1 | CTD_human | |
| Tgene | C0233796 | Temporary Amnesia | 1 | CTD_human | |
| Tgene | C0236795 | Dissociative Amnesia | 1 | CTD_human | |
| Tgene | C0262497 | Global Amnesia | 1 | CTD_human | |
| Tgene | C0750906 | Tactile Amnesia | 1 | CTD_human | |
| Tgene | C0750907 | Amnestic State | 1 | CTD_human | |
| Tgene | C1865295 | Auriculo-condylar syndrome | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C1876185 | Dysgnathia complex | 1 | GENOMICS_ENGLAND |
