
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:AGK-BRAF (FusionGDB2 ID:HG55750TG673) |
Fusion Gene Summary for AGK-BRAF |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: AGK-BRAF | Fusion gene ID: hg55750tg673 | Hgene | Tgene | Gene symbol | AGK | BRAF | Gene ID | 55750 | 673 |
| Gene name | acylglycerol kinase | B-Raf proto-oncogene, serine/threonine kinase | |
| Synonyms | CATC5|CTRCT38|MTDPS10|MULK | B-RAF1|B-raf|BRAF1|NS7|RAFB1 | |
| Cytomap | ('AGK')('BRAF') 7q34 | 7q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | acylglycerol kinase, mitochondrialhAGKhsMuLKmulti-substrate lipid kinasemultiple substrate lipid kinase | serine/threonine-protein kinase B-raf94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)B-Raf serine/threonine-proteinmurine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafv-raf murine sarcoma viral oncogene | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | P15056 | |
| Ensembl transtripts involved in fusion gene | ENST00000495028, ENST00000473247, ENST00000355413, ENST00000492693, ENST00000535825, | ENST00000492693, ENST00000495028, ENST00000535825, ENST00000355413, ENST00000473247, | |
| Fusion gene scores | * DoF score | 6 X 5 X 6=180 | 48 X 58 X 16=44544 |
| # samples | 8 | 69 | |
| ** MAII score | log2(8/180*10)=-1.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(69/44544*10)=-6.0124909441832 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: AGK [Title/Abstract] AND BRAF [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | AGK(141255367)-BRAF(140494267), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BRAF-AGK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BRAF-AGK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. AGK-BRAF seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. AGK-BRAF seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. AGK-BRAF seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. AGK-BRAF seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AGK | GO:0045039 | protein import into mitochondrial inner membrane | 28712724|28712726 |
| Tgene | BRAF | GO:0000186 | activation of MAPKK activity | 29433126 |
| Tgene | BRAF | GO:0006468 | protein phosphorylation | 17563371 |
| Tgene | BRAF | GO:0010828 | positive regulation of glucose transmembrane transport | 23010278 |
| Tgene | BRAF | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 |
| Tgene | BRAF | GO:0043066 | negative regulation of apoptotic process | 19667065 |
| Tgene | BRAF | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 |
| Tgene | BRAF | GO:0071277 | cellular response to calcium ion | 18567582 |
| Tgene | BRAF | GO:0090150 | establishment of protein localization to membrane | 23010278 |
 Fusion gene breakpoints across AGK (5'-gene) Fusion gene breakpoints across AGK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
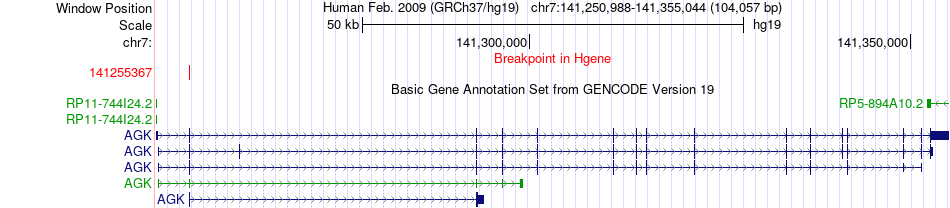 |
 Fusion gene breakpoints across BRAF (3'-gene) Fusion gene breakpoints across BRAF (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
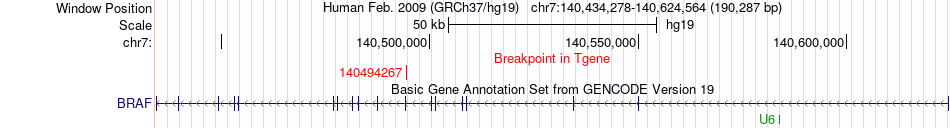 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-DA-A1IA-06A | AGK | chr7 | 141255367 | - | BRAF | chr7 | 140494267 | - |
| ChimerDB4 | SKCM | TCGA-DA-A1IA-06A | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| ChimerDB4 | THCA | TCGA-DJ-A2PX-01A | AGK | chr7 | 141255367 | - | BRAF | chr7 | 140494267 | - |
| ChimerKB4 | . | . | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
Top |
Fusion Gene ORF analysis for AGK-BRAF |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000495028 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| Frame-shift | ENST00000473247 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| In-frame | ENST00000355413 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| In-frame | ENST00000492693 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| In-frame | ENST00000535825 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - |
| intron-intron | ENST00000355413 | ENST00000288602 | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
| intron-intron | ENST00000473247 | ENST00000288602 | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
| intron-intron | ENST00000492693 | ENST00000288602 | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
| intron-intron | ENST00000495028 | ENST00000288602 | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
| intron-intron | ENST00000535825 | ENST00000288602 | AGK | chr7 | 141255252 | + | BRAF | chr7 | 141255252 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000355413 | AGK | chr7 | 141255367 | + | ENST00000288602 | BRAF | chr7 | 140494267 | - | 1800 | 361 | 260 | 1681 | 473 |
| ENST00000535825 | AGK | chr7 | 141255367 | + | ENST00000288602 | BRAF | chr7 | 140494267 | - | 1579 | 140 | 39 | 1460 | 473 |
| ENST00000492693 | AGK | chr7 | 141255367 | + | ENST00000288602 | BRAF | chr7 | 140494267 | - | 1554 | 115 | 14 | 1435 | 473 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000355413 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - | 0.005953697 | 0.99404633 |
| ENST00000535825 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - | 0.003412508 | 0.9965875 |
| ENST00000492693 | ENST00000288602 | AGK | chr7 | 141255367 | + | BRAF | chr7 | 140494267 | - | 0.002883715 | 0.9971163 |
Top |
Fusion Genomic Features for AGK-BRAF |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
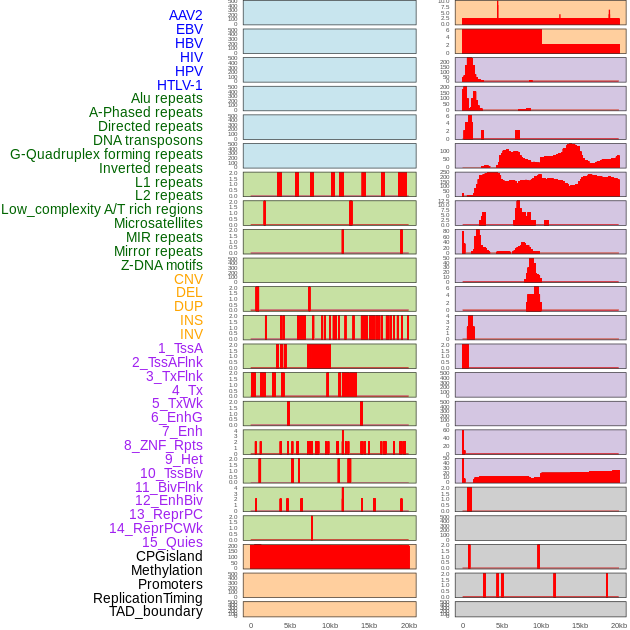 |
Top |
Fusion Protein Features for AGK-BRAF |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:141255367/chr7:140494267) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | BRAF |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGK | chr7:141255367 | chr7:140494267 | ENST00000355413 | + | 2 | 16 | 15_31 | 33 | 423.0 | Region | Hydrophobic |
| Hgene | AGK | chr7:141255367 | chr7:140494267 | ENST00000492693 | + | 1 | 3 | 15_31 | 33 | 66.0 | Region | Hydrophobic |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 428_432 | 326 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 457_717 | 326 | 767.0 | Domain | Protein kinase | |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 463_471 | 326 | 767.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGK | chr7:141255367 | chr7:140494267 | ENST00000355413 | + | 2 | 16 | 58_199 | 33 | 423.0 | Domain | DAGKc |
| Hgene | AGK | chr7:141255367 | chr7:140494267 | ENST00000492693 | + | 1 | 3 | 58_199 | 33 | 66.0 | Domain | DAGKc |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 122_129 | 326 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 6_11 | 326 | 767.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 155_227 | 326 | 767.0 | Domain | RBD | |
| Tgene | BRAF | chr7:141255367 | chr7:140494267 | ENST00000288602 | 6 | 18 | 234_280 | 326 | 767.0 | Zinc finger | Phorbol-ester/DAG-type |
Top |
Fusion Gene Sequence for AGK-BRAF |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >2885_2885_1_AGK-BRAF_AGK_chr7_141255367_ENST00000355413_BRAF_chr7_140494267_ENST00000288602_length(transcript)=1800nt_BP=361nt ATGAAAGGAAGAGTGAGCCAATAAACGAGGGAACGCCGGGAAAGGCAGCCTCAAGCCGGTGGGCCCTGCCACCCCCACCGTCCCTGAGCA TCCGAGCCGGTTCCCGCCCCCGGCCCGAACTGGCCCGCGCGCGCTCGCAGCCCCGCGGCGGAACCCGAGGGCGGCGGCAGCGGTTTCCCT TGAACGAGCCGGGGAATCTGGAGGGAGCACACAGGAAAGGCAGAGCCGCGAGCTGGACCAGCCGTGCAAATCTCTAGAAGATGACGGTGT TCTTTAAAACGCTTCGAAATCACTGGAAGAAAACTACAGCTGGGCTCTGCCTGCTGACCTGGGGAGGCCATTGGCTCTATGGAAAACACT GGCCCCAAATTCTCACCAGTCCGTCTCCTTCAAAATCCATTCCAATTCCACAGCCCTTCCGACCAGCAGATGAAGATCATCGAAATCAAT TTGGGCAACGAGACCGATCCTCATCAGCTCCCAATGTGCATATAAACACAATAGAACCTGTCAATATTGATGACTTGATTAGAGACCAAG GATTTCGTGGTGATGGAGGATCAACCACAGGTTTGTCTGCTACCCCCCCTGCCTCATTACCTGGCTCACTAACTAACGTGAAAGCCTTAC AGAAATCTCCAGGACCTCAGCGAGAAAGGAAGTCATCTTCATCCTCAGAAGACAGGAATCGAATGAAAACACTTGGTAGACGGGACTCGA GTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTGGATCTGGATCATTTGGAACAGTCTACAAGGGAAAGTGGC ATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCTACACCTCAGCAGTTACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGA AAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCCACAAAGCCACAACTGGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCT TGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATGATCAAACTTATAGATATTGCACGACAGACTGCACAGGGCATGGATTACT TACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTC TAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTGGATCCATTTTGTGGATGGCACCAGAAGTCATCAGAA TGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATATGCATTTGGAATTGTTCTGTATGAATTGATGACTGGACAGTTACCTTATT CAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGACGAGGATACCTGTCTCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAA AAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAAAGAGATGAGAGACCACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGC TGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAACCCTCCTTGAATCGGGCTGGTTTCCAAACAGAGGATTTTAGTCTATATG CTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATATGGTGCGTTTCCTGTCCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAG TAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATATGTTAAATTGAATAAAATACTCTCTTTTTTTTTAAGGTGAACCAAAGAA >2885_2885_1_AGK-BRAF_AGK_chr7_141255367_ENST00000355413_BRAF_chr7_140494267_ENST00000288602_length(amino acids)=473AA_BP=34 MTVFFKTLRNHWKKTTAGLCLLTWGGHWLYGKHWPQILTSPSPSKSIPIPQPFRPADEDHRNQFGQRDRSSSAPNVHINTIEPVNIDDLI RDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSEDRNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYK GKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQG MDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQ LPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHRSASEPSLNRAGFQTEDF SLYACASPKTPIQAGGYGAFPVH -------------------------------------------------------------- >2885_2885_2_AGK-BRAF_AGK_chr7_141255367_ENST00000492693_BRAF_chr7_140494267_ENST00000288602_length(transcript)=1554nt_BP=115nt CAAATCTCTAGAAGATGACGGTGTTCTTTAAAACGCTTCGAAATCACTGGAAGAAAACTACAGCTGGGCTCTGCCTGCTGACCTGGGGAG GCCATTGGCTCTATGGAAAACACTGGCCCCAAATTCTCACCAGTCCGTCTCCTTCAAAATCCATTCCAATTCCACAGCCCTTCCGACCAG CAGATGAAGATCATCGAAATCAATTTGGGCAACGAGACCGATCCTCATCAGCTCCCAATGTGCATATAAACACAATAGAACCTGTCAATA TTGATGACTTGATTAGAGACCAAGGATTTCGTGGTGATGGAGGATCAACCACAGGTTTGTCTGCTACCCCCCCTGCCTCATTACCTGGCT CACTAACTAACGTGAAAGCCTTACAGAAATCTCCAGGACCTCAGCGAGAAAGGAAGTCATCTTCATCCTCAGAAGACAGGAATCGAATGA AAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGACAAAGAATTGGATCTGGATCATTTG GAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCTACACCTCAGCAGTTACAAGCCTTCA AAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCCACAAAGCCACAACTGGCTATTGTTA CCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATGATCAAACTTATAGATATTGCACGAC AGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTCATGAAGACCTCA CAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTGGATCCATTTTGT GGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATATGCATTTGGAATTGTTCTGTATGAAT TGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGACGAGGATACCTGTCTCCAGATCTCA GTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAAAGAGATGAGAGACCACTCTTTCCCC AAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAACCCTCCTTGAATCGGGCTGGTTTCC AAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATATGGTGCGTTTCCTGTCCACTGAAACA AATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATATGTTAAATTGAATAAAATACTCTCT TTTTTTTTAAGGTGAACCAAAGAA >2885_2885_2_AGK-BRAF_AGK_chr7_141255367_ENST00000492693_BRAF_chr7_140494267_ENST00000288602_length(amino acids)=473AA_BP=34 MTVFFKTLRNHWKKTTAGLCLLTWGGHWLYGKHWPQILTSPSPSKSIPIPQPFRPADEDHRNQFGQRDRSSSAPNVHINTIEPVNIDDLI RDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSEDRNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYK GKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQG MDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQ LPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHRSASEPSLNRAGFQTEDF SLYACASPKTPIQAGGYGAFPVH -------------------------------------------------------------- >2885_2885_3_AGK-BRAF_AGK_chr7_141255367_ENST00000535825_BRAF_chr7_140494267_ENST00000288602_length(transcript)=1579nt_BP=140nt AGAGCCGCGAGCTGGACCAGCCGTGCAAATCTCTAGAAGATGACGGTGTTCTTTAAAACGCTTCGAAATCACTGGAAGAAAACTACAGCT GGGCTCTGCCTGCTGACCTGGGGAGGCCATTGGCTCTATGGAAAACACTGGCCCCAAATTCTCACCAGTCCGTCTCCTTCAAAATCCATT CCAATTCCACAGCCCTTCCGACCAGCAGATGAAGATCATCGAAATCAATTTGGGCAACGAGACCGATCCTCATCAGCTCCCAATGTGCAT ATAAACACAATAGAACCTGTCAATATTGATGACTTGATTAGAGACCAAGGATTTCGTGGTGATGGAGGATCAACCACAGGTTTGTCTGCT ACCCCCCCTGCCTCATTACCTGGCTCACTAACTAACGTGAAAGCCTTACAGAAATCTCCAGGACCTCAGCGAGAAAGGAAGTCATCTTCA TCCTCAGAAGACAGGAATCGAATGAAAACACTTGGTAGACGGGACTCGAGTGATGATTGGGAGATTCCTGATGGGCAGATTACAGTGGGA CAAAGAATTGGATCTGGATCATTTGGAACAGTCTACAAGGGAAAGTGGCATGGTGATGTGGCAGTGAAAATGTTGAATGTGACAGCACCT ACACCTCAGCAGTTACAAGCCTTCAAAAATGAAGTAGGAGTACTCAGGAAAACACGACATGTGAATATCCTACTCTTCATGGGCTATTCC ACAAAGCCACAACTGGCTATTGTTACCCAGTGGTGTGAGGGCTCCAGCTTGTATCACCATCTCCATATCATTGAGACCAAATTTGAGATG ATCAAACTTATAGATATTGCACGACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAAT AATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGTGAAATCTCGATGGAGTGGGTCCCATCAGTTT GAACAGTTGTCTGGATCCATTTTGTGGATGGCACCAGAAGTCATCAGAATGCAAGATAAAAATCCATACAGCTTTCAGTCAGATGTATAT GCATTTGGAATTGTTCTGTATGAATTGATGACTGGACAGTTACCTTATTCAAACATCAACAACAGGGACCAGATAATTTTTATGGTGGGA CGAGGATACCTGTCTCCAGATCTCAGTAAGGTACGGAGTAACTGTCCAAAAGCCATGAAGAGATTAATGGCAGAGTGCCTCAAAAAGAAA AGAGATGAGAGACCACTCTTTCCCCAAATTCTCGCCTCTATTGAGCTGCTGGCCCGCTCATTGCCAAAAATTCACCGCAGTGCATCAGAA CCCTCCTTGAATCGGGCTGGTTTCCAAACAGAGGATTTTAGTCTATATGCTTGTGCTTCTCCAAAAACACCCATCCAGGCAGGGGGATAT GGTGCGTTTCCTGTCCACTGAAACAAATGAGTGAGAGAGTTCAGGAGAGTAGCAACAAAAGGAAAATAAATGAACATATGTTTGCTTATA TGTTAAATTGAATAAAATACTCTCTTTTTTTTTAAGGTGAACCAAAGAA >2885_2885_3_AGK-BRAF_AGK_chr7_141255367_ENST00000535825_BRAF_chr7_140494267_ENST00000288602_length(amino acids)=473AA_BP=34 MTVFFKTLRNHWKKTTAGLCLLTWGGHWLYGKHWPQILTSPSPSKSIPIPQPFRPADEDHRNQFGQRDRSSSAPNVHINTIEPVNIDDLI RDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSPGPQRERKSSSSSEDRNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYK GKWHGDVAVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHHLHIIETKFEMIKLIDIARQTAQG MDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATVKSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQ LPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHRSASEPSLNRAGFQTEDF SLYACASPKTPIQAGGYGAFPVH -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for AGK-BRAF |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for AGK-BRAF |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Tgene | BRAF | P15056 | DB08881 | Vemurafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB08881 | Vemurafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB08881 | Vemurafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB08896 | Regorafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB08896 | Regorafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB08896 | Regorafenib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB14840 | Ripretinib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB14840 | Ripretinib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB14840 | Ripretinib | Inhibitor | Small molecule | Approved |
| Tgene | BRAF | P15056 | DB00398 | Sorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB00398 | Sorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB00398 | Sorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB08912 | Dabrafenib | Antagonist|Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB08912 | Dabrafenib | Antagonist|Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB08912 | Dabrafenib | Antagonist|Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB11718 | Encorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB11718 | Encorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB11718 | Encorafenib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB12010 | Fostamatinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB12010 | Fostamatinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | BRAF | P15056 | DB12010 | Fostamatinib | Inhibitor | Small molecule | Approved|Investigational |
Top |
Related Diseases for AGK-BRAF |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | AGK | C1859317 | Cataract and cardiomyopathy | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | AGK | C0266539 | Congenital total cataract | 2 | ORPHANET |
| Hgene | AGK | C3553494 | CATARACT 38 | 1 | CTD_human;GENOMICS_ENGLAND |
| Tgene | C0025202 | melanoma | 24 | CGI;CTD_human;UNIPROT | |
| Tgene | C1275081 | Cardio-facio-cutaneous syndrome | 14 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0009402 | Colorectal Carcinoma | 8 | CTD_human;UNIPROT | |
| Tgene | C0028326 | Noonan Syndrome | 8 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C0238463 | Papillary thyroid carcinoma | 8 | CTD_human;ORPHANET | |
| Tgene | C0040136 | Thyroid Neoplasm | 6 | CGI;CTD_human | |
| Tgene | C0151468 | Thyroid Gland Follicular Adenoma | 6 | CTD_human | |
| Tgene | C0175704 | LEOPARD Syndrome | 6 | CLINGEN;GENOMICS_ENGLAND | |
| Tgene | C0549473 | Thyroid carcinoma | 6 | CGI;CTD_human | |
| Tgene | C3150970 | NOONAN SYNDROME 7 | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0009404 | Colorectal Neoplasms | 4 | CTD_human | |
| Tgene | C3150971 | LEOPARD SYNDROME 3 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C1519086 | Pilomyxoid astrocytoma | 3 | ORPHANET | |
| Tgene | C0004565 | Melanoma, B16 | 2 | CTD_human | |
| Tgene | C0009075 | Melanoma, Cloudman S91 | 2 | CTD_human | |
| Tgene | C0018598 | Melanoma, Harding-Passey | 2 | CTD_human | |
| Tgene | C0023443 | Hairy Cell Leukemia | 2 | CGI;ORPHANET | |
| Tgene | C0025205 | Melanoma, Experimental | 2 | CTD_human | |
| Tgene | C0033578 | Prostatic Neoplasms | 2 | CTD_human | |
| Tgene | C0152013 | Adenocarcinoma of lung (disorder) | 2 | CGI;CTD_human | |
| Tgene | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human | |
| Tgene | C0587248 | Costello syndrome (disorder) | 2 | CLINGEN;CTD_human | |
| Tgene | C3501843 | Nonmedullary Thyroid Carcinoma | 2 | CTD_human | |
| Tgene | C3501844 | Familial Nonmedullary Thyroid Cancer | 2 | CTD_human | |
| Tgene | C0002448 | Ameloblastoma | 1 | CTD_human | |
| Tgene | C0004114 | Astrocytoma | 1 | CTD_human | |
| Tgene | C0010276 | Craniopharyngioma | 1 | CTD_human;ORPHANET | |
| Tgene | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human | |
| Tgene | C0017638 | Glioma | 1 | CGI;CTD_human | |
| Tgene | C0019621 | Histiocytosis, Langerhans-Cell | 1 | CGI;ORPHANET | |
| Tgene | C0022665 | Kidney Neoplasm | 1 | CTD_human | |
| Tgene | C0023903 | Liver neoplasms | 1 | CTD_human | |
| Tgene | C0024232 | Lymphatic Metastasis | 1 | CTD_human | |
| Tgene | C0024694 | Mandibular Neoplasms | 1 | CTD_human | |
| Tgene | C0027659 | Neoplasms, Experimental | 1 | CTD_human | |
| Tgene | C0027962 | Melanocytic nevus | 1 | GENOMICS_ENGLAND | |
| Tgene | C0036920 | Sezary Syndrome | 1 | CTD_human | |
| Tgene | C0041409 | Turner Syndrome, Male | 1 | CTD_human | |
| Tgene | C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | CTD_human | |
| Tgene | C0205768 | Subependymal Giant Cell Astrocytoma | 1 | CTD_human | |
| Tgene | C0206686 | Adrenocortical carcinoma | 1 | CTD_human | |
| Tgene | C0206754 | Neuroendocrine Tumors | 1 | CTD_human | |
| Tgene | C0259783 | mixed gliomas | 1 | CTD_human | |
| Tgene | C0278875 | Adult Craniopharyngioma | 1 | CTD_human | |
| Tgene | C0280783 | Juvenile Pilocytic Astrocytoma | 1 | CTD_human | |
| Tgene | C0280785 | Diffuse Astrocytoma | 1 | CTD_human | |
| Tgene | C0334579 | Anaplastic astrocytoma | 1 | CGI;CTD_human | |
| Tgene | C0334580 | Protoplasmic astrocytoma | 1 | CTD_human | |
| Tgene | C0334581 | Gemistocytic astrocytoma | 1 | CTD_human | |
| Tgene | C0334582 | Fibrillary Astrocytoma | 1 | CTD_human | |
| Tgene | C0334583 | Pilocytic Astrocytoma | 1 | CGI;CTD_human | |
| Tgene | C0338070 | Childhood Cerebral Astrocytoma | 1 | CTD_human | |
| Tgene | C0345904 | Malignant neoplasm of liver | 1 | CTD_human | |
| Tgene | C0376407 | Granulomatous Slack Skin | 1 | CTD_human | |
| Tgene | C0406803 | Syringocystadenoma Papilliferum | 1 | GENOMICS_ENGLAND | |
| Tgene | C0431128 | Papillary craniopharyngioma | 1 | CTD_human | |
| Tgene | C0431129 | Adamantinous Craniopharyngioma | 1 | CTD_human | |
| Tgene | C0547065 | Mixed oligoastrocytoma | 1 | CTD_human | |
| Tgene | C0555198 | Malignant Glioma | 1 | CTD_human | |
| Tgene | C0596263 | Carcinogenesis | 1 | CTD_human | |
| Tgene | C0684249 | Carcinoma of lung | 1 | CGI;UNIPROT | |
| Tgene | C0740457 | Malignant neoplasm of kidney | 1 | CTD_human | |
| Tgene | C0750935 | Cerebral Astrocytoma | 1 | CTD_human | |
| Tgene | C0750936 | Intracranial Astrocytoma | 1 | CTD_human | |
| Tgene | C0751061 | Craniopharyngioma, Child | 1 | CTD_human | |
| Tgene | C0920269 | Microsatellite Instability | 1 | CTD_human | |
| Tgene | C1527404 | Female Pseudo-Turner Syndrome | 1 | CTD_human | |
| Tgene | C1704230 | Grade I Astrocytoma | 1 | CTD_human | |
| Tgene | C1721098 | Replication Error Phenotype | 1 | CTD_human | |
| Tgene | C2239176 | Liver carcinoma | 1 | CTD_human | |
| Tgene | C4551484 | Leopard Syndrome 1 | 1 | GENOMICS_ENGLAND | |
| Tgene | C4551602 | Noonan Syndrome 1 | 1 | CTD_human | |
| Tgene | C4721532 | Lymphoma, Non-Hodgkin, Familial | 1 | UNIPROT | |
| Tgene | C4733333 | familial non-medullary thyroid cancer | 1 | GENOMICS_ENGLAND |
