
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ARID1B-NKAIN2 (FusionGDB2 ID:HG57492TG154215) |
Fusion Gene Summary for ARID1B-NKAIN2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ARID1B-NKAIN2 | Fusion gene ID: hg57492tg154215 | Hgene | Tgene | Gene symbol | ARID1B | NKAIN2 | Gene ID | 57492 | 154215 |
| Gene name | AT-rich interaction domain 1B | sodium/potassium transporting ATPase interacting 2 | |
| Synonyms | 6A3-5|BAF250B|BRIGHT|CSS1|DAN15|ELD/OSA1|MRD12|OSA2|P250R | FAM77B|NKAIP2|TCBA|TCBA1 | |
| Cytomap | ('ARID1B')('NKAIN2') 6q25.3 | 6q22.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | AT-rich interactive domain-containing protein 1BARID domain-containing protein 1BAT rich interactive domain 1B (SWI1-like)BRG1-associated factor 250bBRG1-binding protein ELD/OSA1ELD (eyelid)/OSA protein | sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2Na(+)/K(+)-transporting ATPase subunit beta-1-interacting protein 2Na+/K+ transporting ATPase interacting 2T-cell lymphoma breakpoint-associated target protein 1 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000275248, ENST00000346085, ENST00000350026, ENST00000367148, ENST00000478761, | ||
| Fusion gene scores | * DoF score | 27 X 18 X 14=6804 | 14 X 12 X 7=1176 |
| # samples | 31 | 18 | |
| ** MAII score | log2(31/6804*10)=-4.45604302038915 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/1176*10)=-2.70781924850669 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ARID1B [Title/Abstract] AND NKAIN2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ARID1B(157256710)-NKAIN2(124979332), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. ARID1B-NKAIN2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ARID1B (5'-gene) Fusion gene breakpoints across ARID1B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
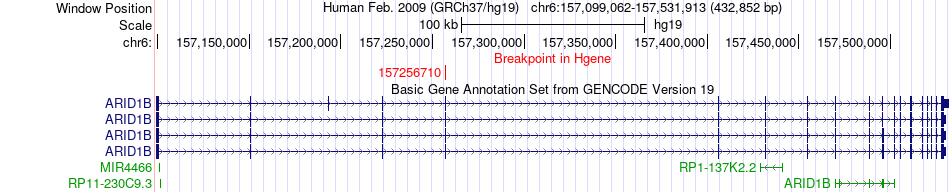 |
 Fusion gene breakpoints across NKAIN2 (3'-gene) Fusion gene breakpoints across NKAIN2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
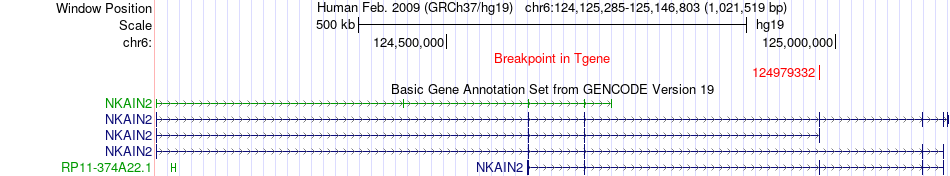 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E2-A1LK-01A | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
Top |
Fusion Gene ORF analysis for ARID1B-NKAIN2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000275248 | ENST00000476571 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000275248 | ENST00000545433 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000275248 | ENST00000546092 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000346085 | ENST00000476571 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000346085 | ENST00000545433 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000346085 | ENST00000546092 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000350026 | ENST00000476571 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000350026 | ENST00000545433 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000350026 | ENST00000546092 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000367148 | ENST00000476571 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000367148 | ENST00000545433 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| 5CDS-intron | ENST00000367148 | ENST00000546092 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| Frame-shift | ENST00000275248 | ENST00000368416 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| Frame-shift | ENST00000346085 | ENST00000368416 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| Frame-shift | ENST00000350026 | ENST00000368416 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| Frame-shift | ENST00000367148 | ENST00000368416 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| In-frame | ENST00000275248 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| In-frame | ENST00000346085 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| In-frame | ENST00000350026 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| In-frame | ENST00000367148 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| intron-3CDS | ENST00000478761 | ENST00000368416 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| intron-3CDS | ENST00000478761 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| intron-intron | ENST00000478761 | ENST00000476571 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| intron-intron | ENST00000478761 | ENST00000545433 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
| intron-intron | ENST00000478761 | ENST00000546092 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000350026 | ARID1B | chr6 | 157256710 | + | ENST00000368417 | NKAIN2 | chr6 | 124979332 | + | 4781 | 1999 | 1 | 2352 | 783 |
| ENST00000346085 | ARID1B | chr6 | 157256710 | + | ENST00000368417 | NKAIN2 | chr6 | 124979332 | + | 4820 | 2038 | 1 | 2391 | 796 |
| ENST00000367148 | ARID1B | chr6 | 157256710 | + | ENST00000368417 | NKAIN2 | chr6 | 124979332 | + | 4780 | 1998 | 0 | 2351 | 783 |
| ENST00000275248 | ARID1B | chr6 | 157256710 | + | ENST00000368417 | NKAIN2 | chr6 | 124979332 | + | 4758 | 1976 | 77 | 2329 | 750 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000350026 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + | 0.008228126 | 0.9917719 |
| ENST00000346085 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + | 0.007764503 | 0.9922355 |
| ENST00000367148 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + | 0.00822867 | 0.9917713 |
| ENST00000275248 | ENST00000368417 | ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979332 | + | 0.009192704 | 0.99080735 |
Top |
Fusion Genomic Features for ARID1B-NKAIN2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979331 | + | 4.77E-06 | 0.99999523 |
| ARID1B | chr6 | 157256710 | + | NKAIN2 | chr6 | 124979331 | + | 4.77E-06 | 0.99999523 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
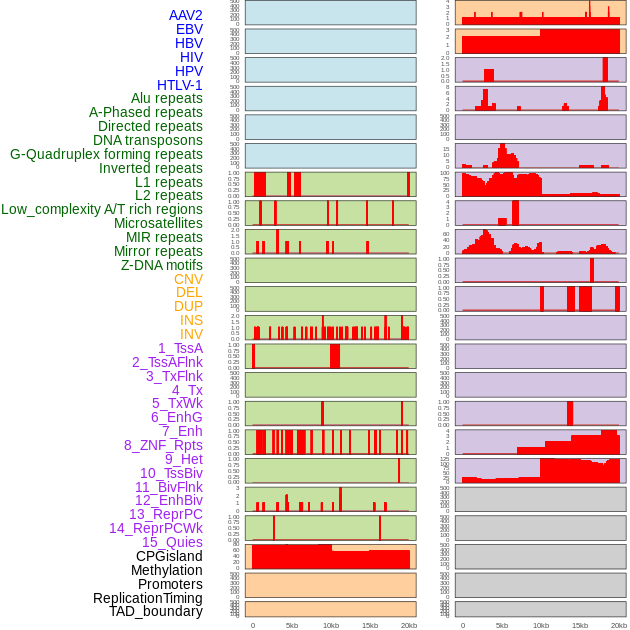 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
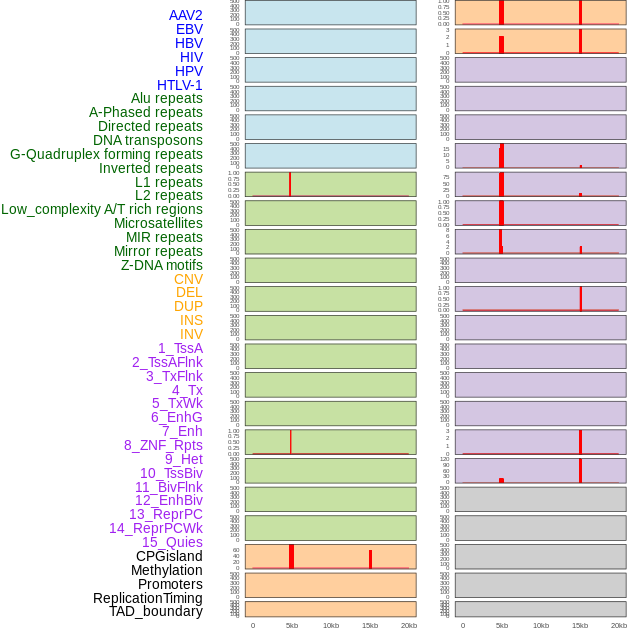 |
Top |
Fusion Protein Features for ARID1B-NKAIN2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:157256710/chr6:124979332) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 107_131 | 679 | 2250.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 114_131 | 679 | 2250.0 | Compositional bias | Note=Poly-Gln |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 141_401 | 679 | 2250.0 | Compositional bias | Note=Gly-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 2_47 | 679 | 2250.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 329_493 | 679 | 2250.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 35_57 | 679 | 2250.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 574_633 | 679 | 2250.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 81_104 | 679 | 2250.0 | Compositional bias | Note=His-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 107_131 | 666 | 2237.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 114_131 | 666 | 2237.0 | Compositional bias | Note=Poly-Gln |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 141_401 | 666 | 2237.0 | Compositional bias | Note=Gly-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 2_47 | 666 | 2237.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 329_493 | 666 | 2237.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 35_57 | 666 | 2237.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 574_633 | 666 | 2237.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 81_104 | 666 | 2237.0 | Compositional bias | Note=His-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 107_131 | 666 | 2290.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 114_131 | 666 | 2290.0 | Compositional bias | Note=Poly-Gln |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 141_401 | 666 | 2290.0 | Compositional bias | Note=Gly-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 2_47 | 666 | 2290.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 329_493 | 666 | 2290.0 | Compositional bias | Note=Ala-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 35_57 | 666 | 2290.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 574_633 | 666 | 2290.0 | Compositional bias | Note=Gln-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 81_104 | 666 | 2290.0 | Compositional bias | Note=His-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 419_423 | 679 | 2250.0 | Motif | Note=LXXLL |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 419_423 | 666 | 2237.0 | Motif | Note=LXXLL |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 419_423 | 666 | 2290.0 | Motif | Note=LXXLL |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368416 | 2 | 4 | 153_173 | 91 | 189.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368417 | 2 | 7 | 153_173 | 91 | 209.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1034_1037 | 679 | 2250.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1441_1444 | 679 | 2250.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1459_1597 | 679 | 2250.0 | Compositional bias | Note=Pro-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1833_1836 | 679 | 2250.0 | Compositional bias | Note=Poly-Pro |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 684_771 | 679 | 2250.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 932_935 | 679 | 2250.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1034_1037 | 666 | 2237.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1441_1444 | 666 | 2237.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1459_1597 | 666 | 2237.0 | Compositional bias | Note=Pro-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1833_1836 | 666 | 2237.0 | Compositional bias | Note=Poly-Pro |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 684_771 | 666 | 2237.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 932_935 | 666 | 2237.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1034_1037 | 666 | 2290.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1441_1444 | 666 | 2290.0 | Compositional bias | Note=Poly-Ser |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1459_1597 | 666 | 2290.0 | Compositional bias | Note=Pro-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1833_1836 | 666 | 2290.0 | Compositional bias | Note=Poly-Pro |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 684_771 | 666 | 2290.0 | Compositional bias | Note=Ser-rich |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 932_935 | 666 | 2290.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1053_1144 | 679 | 2250.0 | Domain | ARID |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1053_1144 | 666 | 2237.0 | Domain | ARID |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1053_1144 | 666 | 2290.0 | Domain | ARID |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 1358_1377 | 679 | 2250.0 | Motif | Nuclear localization signal |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000346085 | + | 5 | 20 | 2036_2040 | 679 | 2250.0 | Motif | Note=LXXLL |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 1358_1377 | 666 | 2237.0 | Motif | Nuclear localization signal |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000350026 | + | 4 | 19 | 2036_2040 | 666 | 2237.0 | Motif | Note=LXXLL |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 1358_1377 | 666 | 2290.0 | Motif | Nuclear localization signal |
| Hgene | ARID1B | chr6:157256710 | chr6:124979332 | ENST00000367148 | + | 4 | 20 | 2036_2040 | 666 | 2290.0 | Motif | Note=LXXLL |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368416 | 2 | 4 | 1_23 | 91 | 189.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368416 | 2 | 4 | 35_55 | 91 | 189.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368416 | 2 | 4 | 62_82 | 91 | 189.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368417 | 2 | 7 | 1_23 | 91 | 209.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368417 | 2 | 7 | 35_55 | 91 | 209.0 | Transmembrane | Helical | |
| Tgene | NKAIN2 | chr6:157256710 | chr6:124979332 | ENST00000368417 | 2 | 7 | 62_82 | 91 | 209.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for ARID1B-NKAIN2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >6409_6409_1_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000275248_NKAIN2_chr6_124979332_ENST00000368417_length(transcript)=4758nt_BP=1976nt CGGCCGCCGCCGGCACCCACAGCGCCAAGAGCGGCGGCTCCGAGGCGGCTCTCAAGGAGGGTGGAAGCGCCGCCGCGCTGTCCTCCTCCT CCTCCTCCTCCGCGGCGGCAGCGGCGGCATCCTCTTCCTCCTCGTCGGGCCCGGGCTCGGCCATGGAGACGGGGCTGCTCCCCAACCACA AACTGAAAACCGTTGGCGAAGCCCCCGCCGCGCCGCCCCACCAGCAGCACCACCACCACCACCATGCCCACCACCACCACCACCATGCCC ACCACCTCCACCACCACCACGCACTACAGCAGCAGCTAAACCAGTTCCAGCAGCAGCAGCAGCAGCAGCAACAGCAGCAGCAGCAGCAGC AGCAACAGCAACATCCCATTTCCAACAACAACAGCTTGGGCGGCGCGGGCGGCGGCGCGCCTCAGCCCGGCCCCGACATGGAGCAGCCGC AACATGGAGGCGCCAAGGACAGTGCTGCGGGCGGCCAGGCCGACCCCCCGGGCCCGCCGCTGCTGAGCAAGCCGGGCGACGAGGACGACG CGCCGCCCAAGATGGGGGAGCCGGCGGGCGGCCGCTACGAGCACCCGGGCTTGGGCGCCCTGGGCACGCAGCAGCCGCCGGTCGCCGTGC CCGGGGGCGGCGGCGGCCCGGCGGCCGTCCCGGAGTTTAATAATTACTATGGCAGCGCTGCCCCTGCGAGCGGCGGCCCCGGCGGCCGCG CTGGGCCTTGCTTTGATCAACATGGCGGACAACAAAGCCCCGGGATGGGGATGATGCACTCCGCCTCCGCCGCCGCCGCCGGGGCCCCCG GCAGCATGGACCCCCTGCAGAACTCCCACGAAGGGTACCCCAACAGCCAGTGCAACCATTATCCGGGCTACAGCCGGCCCGGCGCGGGCG GCGGCGGCGGCGGCGGCGGCGGAGGAGGAGGAGGCAGCGGAGGAGGAGGAGGAGGAGGAGGAGCAGGAGCAGGAGGAGCAGGAGCGGGAG CTGTGGCGGCGGCGGCCGCGGCGGCGGCGGCAGCAGCAGGAGGCGGCGGCGGCGGCGGCTATGGGGGCTCGTCCGCGGGGTACGGGGTGC TGAGCTCCCCCCGGCAGCAGGGCGGCGGCATGATGATGGGCCCCGGGGGCGGCGGGGCCGCGAGCCTCAGCAAGGCGGCCGCCGGCTCGG CGGCGGGGGGCTTCCAGCGCTTCGCCGGCCAGAACCAGCACCCGTCGGGGGCCACCCCGACCCTCAATCAGCTGCTCACCTCGCCCAGCC CCATGATGCGGAGCTACGGCGGCAGCTACCCCGAGTACAGCAGCCCCAGCGCGCCGCCGCCGCCGCCGTCGCAGCCCCAGTCCCAGGCGG CGGCGGCGGGGGCGGCGGCGGGCGGCCAGCAGGCGGCCGCGGGCATGGGCTTGGGCAAGGACATGGGCGCCCAGTACGCCGCTGCCAGCC CGGCCTGGGCGGCCGCGCAACAAAGGAGTCACCCGGCGATGAGCCCCGGCACCCCCGGACCGACCATGGGCAGATCCCAGGGCAGCCCAA TGGATCCAATGGTGATGAAGAGACCTCAGTTGTATGGCATGGGCAGTAACCCTCATTCTCAGCCTCAGCAGAGCAGTCCGTACCCAGGAG GTTCCTATGGCCCTCCAGGCCCACAGCGGTATCCAATTGGCATCCAGGGTCGGACTCCCGGGGCCATGGCCGGAATGCAGTACCCTCAGC AGCAGATGCCACCTCAGTATGGACAGCAAGGTGTGAGTGGTTACTGCCAGCAGGGCCAACAGCCATATTACAGCCAGCAGCCGCAGCCCC CGCACCTCCCACCCCAGGCGCAGTATCTGCCGTCCCAGTCCCAGCAGAGGTACCAGCCGCAGCAGGACATGTCTCAGGAAGGCTATGGAA CTAGATCTCAACCTCCTCTGGCCCCCGGAAAACCTAACCATGAAGACTTGAACTTAATACAGCAAGAAAGACCATCAAGTTTACCAGAAA CAGACCTTATCCTGACTTTTAATATATCAATGCACCGATCTTGGTGGATGGAGAATGGACCAGGATGTACGGTGACGTCAGTGACACCTG CCCCAGACTGGGCCCCAGAAGACCATCGCTACATCACGGTCTCAGGGTGTTTGCTGGAGTACCAGTACATAGAAGTGGCTCATAGTTCCC TCCAGATTGTCCTCGCACTGGCAGGTTTCATCTACGCCTGTTATGTTGTGAAATGTATAACTGAAGAAGAGGACAGCTTTGATTTCATAG GTGGCTTTGACTCTTATGGCTATCAAGGGCCTCAGAAGACATCTCATTTACAACTACAGCCTATGTACATGTCAAAATAATACAGATGAC TTCAGTATGTCAGCCCATGGACCTTTCAAAGAACTTTTTTCGCAGTGGCCTCCTGCATTTCATGAAGAGCAAGAAGCAACTGAGTTTAAA TACATACACGTATTAACAAAACAAATGCAAAGCCTCTACATACAACACTGACACACACACACACACACACGTGAGCACGCACACACCAAT TCCACTTGACCTCCTCTTTCTAACTGAAACAGACAAATATGCAGGACACGCCCATCTTGGATTTCCTGAAAGCAGGCCCCTTTCTCCCAG GCCTTCGGAAAGTTCAGAAGGAGATGTGTTGATGCCCAACGGTTGCCGGCCATTGCTAACTCCTCTGCAGCCCAGCGGGTTGGCCTCTGT GAGCTGGGAAGTCATCCAAGGCACATTAGTTTGAGAGCTCTGTCTTCTGCACTCCATACATCTTGACAGCACCACGGCTACTTAGGCAAT GTAATTGCAAAAACAAACGAAACATGCCATGATGACCTTGTACCCGAAGTCCGAAATGGCAGATTGTTTGCCCTAAAGGCTGTACCGTAC ATACTTGCCTTAACCCACCTACATTGTGTGCCCAAGAATCCCAGTGCAGACAGCCCCAAGCCACTTCCTTTAGGCTAACACACTGCTATT AATTCTAAATGAGTTCAAGCCTGTGTGTTTCATGTCAAGGAACAAGACTGAATAGCTCAGCAGCTCCAGTGTGTCTGTATAAGAAAAGTG CCAGCTGTCAGCAAAGTGCTTTACCATAGAGCCTCCATGGTGGCCCAAAAGAGACAAGTAACAATAACAATATCTGTCATCTTTATAATT CTAAAATATGGTCTTCTGATGGGGTTGGGGGTGGGACAGAGTAAAACATTAATCTACTAAACTCACTAATCTACTGGAAGCAGAAAGCAG AAATGGCTTAACTCGCTGATTTTATTTATTGCACTCTAGGTGCTTTTATAAGTATCTACAATTGTCTTTTACTAACACAGAAACAGCTGT GGATTTCTATCAACAGAGGCCAACTGAATATGTATTAGCTATGTTTACTCTTTTATATCTAATTCAAATTGAAGACTCTACGTAGGTGAA CTTTTTTTTCTAACTTCAGTGTACAAGATTATTTTTGGGTGGGTGGGGGGAGCAATAGTTACCTTGGTGAAATATTTGAGATCTTTTTGT GGTATTTGGGGGATCTGTTGTGTGTTAGAGTGTATTTCAACCCTCCTGTATTATTGTGGCAGAAAAACTGTGCTGTTAACGTAGTTGGCA ACAAGATGCCTTTGGAAACTTAATAGTAGGTCAATGTGTTTATGGAATATTCTGAACTCCTAGGAAATTTTGTTCTATATTTTAAAGCAT TATTTGGTTTTCATTAGTGGAAAACATAATTAGCTCAGGCTTATCAGGAACTCAGATGAGTCATATTTTTTCTTGGATACACCTGAGACA TTAAACTCAATTTTTTAAAACTAACACACCATTCTTTAGTGCAGTGAAATTCAAAAGTTATAAATTGGCATGGAGCAAGTTTTTTCTTTG ATTTCTATGAAGTTTCAGTTATTGGATTAAACTACTTTAGACCTTAGCCCATTCGGATCTACATACACCAGTTCCATTACTGTGTTTGCA TGGATGTCTGTAATATACACACACATATACATTTGAAAATGATAGAGGAACAAAGAGTTATCATTGGAACATAAACTATAAAAAAAGATT CCAGCACTCATGCTTGAATTTCCTCAGTGGGTTGTATGTTTGTCCATACATACATGTATAAAATAAACAATTGCAAACATGTCAGTGAAA CCTCATATGACGCTTTTATAGTGACATGCATGTTTAATGTATGAGAAAAATATTTACCAATGTATTGTCCTTTGAGTCCTAAAATATAAA TACTATACATGCCAGAAACTATTCTAATGTGACTTTTAATGTGACTATTAAAAATGAGGTTTCACTGTACATTCATGCTGTGATGCGATT TCACCTTATAGTACATGTATATGGGTACAGAAAATAAACCCTCATATTCCTGCTTTAAGTTACAGCCAAGTTTTCACCAAATCACAACTA TCATCACCACCAAAATTTTAAAAAACAGTAGTTGACTACTAAAAAGTAATCAGTGGACAGGTCATTTATGGGATCAGTAAGCACAGTAGT GTGATTATATCTGGGAAAACATCTACAGTTGTACAGCTGTATCCTCCTCAAAATCCGGTGAAGAATGCTATTTTTCTAGGCTGAGCTTTT GTTATAAACTTAATATTCAGAAGGCAAGGGTTATGATCCTGATGTGTCCTTTTTTTTTGCATTTGTTAATTCTGAATGTATTTTTAACAG AATGATTTTTTTGACTTACTAGTGTAATTTGCATTTTAAAAATAAATGGAAAAATACTTAAGTTTCTGAGCCATCCCT >6409_6409_1_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000275248_NKAIN2_chr6_124979332_ENST00000368417_length(amino acids)=750AA_BP=631 MSSSSSSSAAAAAASSSSSSGPGSAMETGLLPNHKLKTVGEAPAAPPHQQHHHHHHAHHHHHHAHHLHHHHALQQQLNQFQQQQQQQQQQ QQQQQQQQHPISNNNSLGGAGGGAPQPGPDMEQPQHGGAKDSAAGGQADPPGPPLLSKPGDEDDAPPKMGEPAGGRYEHPGLGALGTQQP PVAVPGGGGGPAAVPEFNNYYGSAAPASGGPGGRAGPCFDQHGGQQSPGMGMMHSASAAAAGAPGSMDPLQNSHEGYPNSQCNHYPGYSR PGAGGGGGGGGGGGGGSGGGGGGGGAGAGGAGAGAVAAAAAAAAAAAGGGGGGGYGGSSAGYGVLSSPRQQGGGMMMGPGGGGAASLSKA AAGSAAGGFQRFAGQNQHPSGATPTLNQLLTSPSPMMRSYGGSYPEYSSPSAPPPPPSQPQSQAAAAGAAAGGQQAAAGMGLGKDMGAQY AAASPAWAAAQQRSHPAMSPGTPGPTMGRSQGSPMDPMVMKRPQLYGMGSNPHSQPQQSSPYPGGSYGPPGPQRYPIGIQGRTPGAMAGM QYPQQQMPPQYGQQGVSGYCQQGQQPYYSQQPQPPHLPPQAQYLPSQSQQRYQPQQDMSQEGYGTRSQPPLAPGKPNHEDLNLIQQERPS SLPETDLILTFNISMHRSWWMENGPGCTVTSVTPAPDWAPEDHRYITVSGCLLEYQYIEVAHSSLQIVLALAGFIYACYVVKCITEEEDS FDFIGGFDSYGYQGPQKTSHLQLQPMYMSK -------------------------------------------------------------- >6409_6409_2_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000346085_NKAIN2_chr6_124979332_ENST00000368417_length(transcript)=4820nt_BP=2038nt CATGGCCCATAACGCGGGCGCCGCGGCCGCCGCCGGCACCCACAGCGCCAAGAGCGGCGGCTCCGAGGCGGCTCTCAAGGAGGGTGGAAG CGCCGCCGCGCTGTCCTCCTCCTCCTCCTCCTCCGCGGCGGCAGCGGCGGCATCCTCTTCCTCCTCGTCGGGCCCGGGCTCGGCCATGGA GACGGGGCTGCTCCCCAACCACAAACTGAAAACCGTTGGCGAAGCCCCCGCCGCGCCGCCCCACCAGCAGCACCACCACCACCACCATGC CCACCACCACCACCACCATGCCCACCACCTCCACCACCACCACGCACTACAGCAGCAGCTAAACCAGTTCCAGCAGCAGCAGCAGCAGCA GCAACAGCAGCAGCAGCAGCAGCAGCAACAGCAACATCCCATTTCCAACAACAACAGCTTGGGCGGCGCGGGCGGCGGCGCGCCTCAGCC CGGCCCCGACATGGAGCAGCCGCAACATGGAGGCGCCAAGGACAGTGCTGCGGGCGGCCAGGCCGACCCCCCGGGCCCGCCGCTGCTGAG CAAGCCGGGCGACGAGGACGACGCGCCGCCCAAGATGGGGGAGCCGGCGGGCGGCCGCTACGAGCACCCGGGCTTGGGCGCCCTGGGCAC GCAGCAGCCGCCGGTCGCCGTGCCCGGGGGCGGCGGCGGCCCGGCGGCCGTCCCGGAGTTTAATAATTACTATGGCAGCGCTGCCCCTGC GAGCGGCGGCCCCGGCGGCCGCGCTGGGCCTTGCTTTGATCAACATGGCGGACAACAAAGCCCCGGGATGGGGATGATGCACTCCGCCTC CGCCGCCGCCGCCGGGGCCCCCGGCAGCATGGACCCCCTGCAGAACTCCCACGAAGGGTACCCCAACAGCCAGTGCAACCATTATCCGGG CTACAGCCGGCCCGGCGCGGGCGGCGGCGGCGGCGGCGGCGGCGGAGGAGGAGGAGGCAGCGGAGGAGGAGGAGGAGGAGGAGGAGCAGG AGCAGGAGGAGCAGGAGCGGGAGCTGTGGCGGCGGCGGCCGCGGCGGCGGCGGCAGCAGCAGGAGGCGGCGGCGGCGGCGGCTATGGGGG CTCGTCCGCGGGGTACGGGGTGCTGAGCTCCCCCCGGCAGCAGGGCGGCGGCATGATGATGGGCCCCGGGGGCGGCGGGGCCGCGAGCCT CAGCAAGGCGGCCGCCGGCTCGGCGGCGGGGGGCTTCCAGCGCTTCGCCGGCCAGAACCAGCACCCGTCGGGGGCCACCCCGACCCTCAA TCAGCTGCTCACCTCGCCCAGCCCCATGATGCGGAGCTACGGCGGCAGCTACCCCGAGTACAGCAGCCCCAGCGCGCCGCCGCCGCCGCC GTCGCAGCCCCAGTCCCAGGCGGCGGCGGCGGGGGCGGCGGCGGGCGGCCAGCAGGCGGCCGCGGGCATGGGCTTGGGCAAGGACATGGG CGCCCAGTACGCCGCTGCCAGCCCGGCCTGGGCGGCCGCGCAACAAAGGAGTCACCCGGCGATGAGCCCCGGCACCCCCGGACCGACCAT GGGCAGATCCCAGGGCAGCCCAATGGATCCAATGGTGATGAAGAGACCTCAGTTGTATGGCATGGGCAGTAACCCTCATTCTCAGCCTCA GCAGAGCAGTCCGTACCCAGGAGGTTCCTATGGCCCTCCAGGCCCACAGCGGTATCCAATTGGCATCCAGGGTCGGACTCCCGGGGCCAT GGCCGGAATGCAGTACCCTCAGCAGCAGGACTCTGGAGATGCCACATGGAAAGAAACATTCTGGTTGATGCCACCTCAGTATGGACAGCA AGGTGTGAGTGGTTACTGCCAGCAGGGCCAACAGCCATATTACAGCCAGCAGCCGCAGCCCCCGCACCTCCCACCCCAGGCGCAGTATCT GCCGTCCCAGTCCCAGCAGAGGTACCAGCCGCAGCAGGACATGTCTCAGGAAGGCTATGGAACTAGATCTCAACCTCCTCTGGCCCCCGG AAAACCTAACCATGAAGACTTGAACTTAATACAGCAAGAAAGACCATCAAGTTTACCAGAAACAGACCTTATCCTGACTTTTAATATATC AATGCACCGATCTTGGTGGATGGAGAATGGACCAGGATGTACGGTGACGTCAGTGACACCTGCCCCAGACTGGGCCCCAGAAGACCATCG CTACATCACGGTCTCAGGGTGTTTGCTGGAGTACCAGTACATAGAAGTGGCTCATAGTTCCCTCCAGATTGTCCTCGCACTGGCAGGTTT CATCTACGCCTGTTATGTTGTGAAATGTATAACTGAAGAAGAGGACAGCTTTGATTTCATAGGTGGCTTTGACTCTTATGGCTATCAAGG GCCTCAGAAGACATCTCATTTACAACTACAGCCTATGTACATGTCAAAATAATACAGATGACTTCAGTATGTCAGCCCATGGACCTTTCA AAGAACTTTTTTCGCAGTGGCCTCCTGCATTTCATGAAGAGCAAGAAGCAACTGAGTTTAAATACATACACGTATTAACAAAACAAATGC AAAGCCTCTACATACAACACTGACACACACACACACACACACGTGAGCACGCACACACCAATTCCACTTGACCTCCTCTTTCTAACTGAA ACAGACAAATATGCAGGACACGCCCATCTTGGATTTCCTGAAAGCAGGCCCCTTTCTCCCAGGCCTTCGGAAAGTTCAGAAGGAGATGTG TTGATGCCCAACGGTTGCCGGCCATTGCTAACTCCTCTGCAGCCCAGCGGGTTGGCCTCTGTGAGCTGGGAAGTCATCCAAGGCACATTA GTTTGAGAGCTCTGTCTTCTGCACTCCATACATCTTGACAGCACCACGGCTACTTAGGCAATGTAATTGCAAAAACAAACGAAACATGCC ATGATGACCTTGTACCCGAAGTCCGAAATGGCAGATTGTTTGCCCTAAAGGCTGTACCGTACATACTTGCCTTAACCCACCTACATTGTG TGCCCAAGAATCCCAGTGCAGACAGCCCCAAGCCACTTCCTTTAGGCTAACACACTGCTATTAATTCTAAATGAGTTCAAGCCTGTGTGT TTCATGTCAAGGAACAAGACTGAATAGCTCAGCAGCTCCAGTGTGTCTGTATAAGAAAAGTGCCAGCTGTCAGCAAAGTGCTTTACCATA GAGCCTCCATGGTGGCCCAAAAGAGACAAGTAACAATAACAATATCTGTCATCTTTATAATTCTAAAATATGGTCTTCTGATGGGGTTGG GGGTGGGACAGAGTAAAACATTAATCTACTAAACTCACTAATCTACTGGAAGCAGAAAGCAGAAATGGCTTAACTCGCTGATTTTATTTA TTGCACTCTAGGTGCTTTTATAAGTATCTACAATTGTCTTTTACTAACACAGAAACAGCTGTGGATTTCTATCAACAGAGGCCAACTGAA TATGTATTAGCTATGTTTACTCTTTTATATCTAATTCAAATTGAAGACTCTACGTAGGTGAACTTTTTTTTCTAACTTCAGTGTACAAGA TTATTTTTGGGTGGGTGGGGGGAGCAATAGTTACCTTGGTGAAATATTTGAGATCTTTTTGTGGTATTTGGGGGATCTGTTGTGTGTTAG AGTGTATTTCAACCCTCCTGTATTATTGTGGCAGAAAAACTGTGCTGTTAACGTAGTTGGCAACAAGATGCCTTTGGAAACTTAATAGTA GGTCAATGTGTTTATGGAATATTCTGAACTCCTAGGAAATTTTGTTCTATATTTTAAAGCATTATTTGGTTTTCATTAGTGGAAAACATA ATTAGCTCAGGCTTATCAGGAACTCAGATGAGTCATATTTTTTCTTGGATACACCTGAGACATTAAACTCAATTTTTTAAAACTAACACA CCATTCTTTAGTGCAGTGAAATTCAAAAGTTATAAATTGGCATGGAGCAAGTTTTTTCTTTGATTTCTATGAAGTTTCAGTTATTGGATT AAACTACTTTAGACCTTAGCCCATTCGGATCTACATACACCAGTTCCATTACTGTGTTTGCATGGATGTCTGTAATATACACACACATAT ACATTTGAAAATGATAGAGGAACAAAGAGTTATCATTGGAACATAAACTATAAAAAAAGATTCCAGCACTCATGCTTGAATTTCCTCAGT GGGTTGTATGTTTGTCCATACATACATGTATAAAATAAACAATTGCAAACATGTCAGTGAAACCTCATATGACGCTTTTATAGTGACATG CATGTTTAATGTATGAGAAAAATATTTACCAATGTATTGTCCTTTGAGTCCTAAAATATAAATACTATACATGCCAGAAACTATTCTAAT GTGACTTTTAATGTGACTATTAAAAATGAGGTTTCACTGTACATTCATGCTGTGATGCGATTTCACCTTATAGTACATGTATATGGGTAC AGAAAATAAACCCTCATATTCCTGCTTTAAGTTACAGCCAAGTTTTCACCAAATCACAACTATCATCACCACCAAAATTTTAAAAAACAG TAGTTGACTACTAAAAAGTAATCAGTGGACAGGTCATTTATGGGATCAGTAAGCACAGTAGTGTGATTATATCTGGGAAAACATCTACAG TTGTACAGCTGTATCCTCCTCAAAATCCGGTGAAGAATGCTATTTTTCTAGGCTGAGCTTTTGTTATAAACTTAATATTCAGAAGGCAAG GGTTATGATCCTGATGTGTCCTTTTTTTTTGCATTTGTTAATTCTGAATGTATTTTTAACAGAATGATTTTTTTGACTTACTAGTGTAAT TTGCATTTTAAAAATAAATGGAAAAATACTTAAGTTTCTGAGCCATCCCT >6409_6409_2_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000346085_NKAIN2_chr6_124979332_ENST00000368417_length(amino acids)=796AA_BP=677 MAHNAGAAAAAGTHSAKSGGSEAALKEGGSAAALSSSSSSSAAAAAASSSSSSGPGSAMETGLLPNHKLKTVGEAPAAPPHQQHHHHHHA HHHHHHAHHLHHHHALQQQLNQFQQQQQQQQQQQQQQQQQQHPISNNNSLGGAGGGAPQPGPDMEQPQHGGAKDSAAGGQADPPGPPLLS KPGDEDDAPPKMGEPAGGRYEHPGLGALGTQQPPVAVPGGGGGPAAVPEFNNYYGSAAPASGGPGGRAGPCFDQHGGQQSPGMGMMHSAS AAAAGAPGSMDPLQNSHEGYPNSQCNHYPGYSRPGAGGGGGGGGGGGGGSGGGGGGGGAGAGGAGAGAVAAAAAAAAAAAGGGGGGGYGG SSAGYGVLSSPRQQGGGMMMGPGGGGAASLSKAAAGSAAGGFQRFAGQNQHPSGATPTLNQLLTSPSPMMRSYGGSYPEYSSPSAPPPPP SQPQSQAAAAGAAAGGQQAAAGMGLGKDMGAQYAAASPAWAAAQQRSHPAMSPGTPGPTMGRSQGSPMDPMVMKRPQLYGMGSNPHSQPQ QSSPYPGGSYGPPGPQRYPIGIQGRTPGAMAGMQYPQQQDSGDATWKETFWLMPPQYGQQGVSGYCQQGQQPYYSQQPQPPHLPPQAQYL PSQSQQRYQPQQDMSQEGYGTRSQPPLAPGKPNHEDLNLIQQERPSSLPETDLILTFNISMHRSWWMENGPGCTVTSVTPAPDWAPEDHR YITVSGCLLEYQYIEVAHSSLQIVLALAGFIYACYVVKCITEEEDSFDFIGGFDSYGYQGPQKTSHLQLQPMYMSK -------------------------------------------------------------- >6409_6409_3_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000350026_NKAIN2_chr6_124979332_ENST00000368417_length(transcript)=4781nt_BP=1999nt CATGGCCCATAACGCGGGCGCCGCGGCCGCCGCCGGCACCCACAGCGCCAAGAGCGGCGGCTCCGAGGCGGCTCTCAAGGAGGGTGGAAG CGCCGCCGCGCTGTCCTCCTCCTCCTCCTCCTCCGCGGCGGCAGCGGCGGCATCCTCTTCCTCCTCGTCGGGCCCGGGCTCGGCCATGGA GACGGGGCTGCTCCCCAACCACAAACTGAAAACCGTTGGCGAAGCCCCCGCCGCGCCGCCCCACCAGCAGCACCACCACCACCACCATGC CCACCACCACCACCACCATGCCCACCACCTCCACCACCACCACGCACTACAGCAGCAGCTAAACCAGTTCCAGCAGCAGCAGCAGCAGCA GCAACAGCAGCAGCAGCAGCAGCAGCAACAGCAACATCCCATTTCCAACAACAACAGCTTGGGCGGCGCGGGCGGCGGCGCGCCTCAGCC CGGCCCCGACATGGAGCAGCCGCAACATGGAGGCGCCAAGGACAGTGCTGCGGGCGGCCAGGCCGACCCCCCGGGCCCGCCGCTGCTGAG CAAGCCGGGCGACGAGGACGACGCGCCGCCCAAGATGGGGGAGCCGGCGGGCGGCCGCTACGAGCACCCGGGCTTGGGCGCCCTGGGCAC GCAGCAGCCGCCGGTCGCCGTGCCCGGGGGCGGCGGCGGCCCGGCGGCCGTCCCGGAGTTTAATAATTACTATGGCAGCGCTGCCCCTGC GAGCGGCGGCCCCGGCGGCCGCGCTGGGCCTTGCTTTGATCAACATGGCGGACAACAAAGCCCCGGGATGGGGATGATGCACTCCGCCTC CGCCGCCGCCGCCGGGGCCCCCGGCAGCATGGACCCCCTGCAGAACTCCCACGAAGGGTACCCCAACAGCCAGTGCAACCATTATCCGGG CTACAGCCGGCCCGGCGCGGGCGGCGGCGGCGGCGGCGGCGGCGGAGGAGGAGGAGGCAGCGGAGGAGGAGGAGGAGGAGGAGGAGCAGG AGCAGGAGGAGCAGGAGCGGGAGCTGTGGCGGCGGCGGCCGCGGCGGCGGCGGCAGCAGCAGGAGGCGGCGGCGGCGGCGGCTATGGGGG CTCGTCCGCGGGGTACGGGGTGCTGAGCTCCCCCCGGCAGCAGGGCGGCGGCATGATGATGGGCCCCGGGGGCGGCGGGGCCGCGAGCCT CAGCAAGGCGGCCGCCGGCTCGGCGGCGGGGGGCTTCCAGCGCTTCGCCGGCCAGAACCAGCACCCGTCGGGGGCCACCCCGACCCTCAA TCAGCTGCTCACCTCGCCCAGCCCCATGATGCGGAGCTACGGCGGCAGCTACCCCGAGTACAGCAGCCCCAGCGCGCCGCCGCCGCCGCC GTCGCAGCCCCAGTCCCAGGCGGCGGCGGCGGGGGCGGCGGCGGGCGGCCAGCAGGCGGCCGCGGGCATGGGCTTGGGCAAGGACATGGG CGCCCAGTACGCCGCTGCCAGCCCGGCCTGGGCGGCCGCGCAACAAAGGAGTCACCCGGCGATGAGCCCCGGCACCCCCGGACCGACCAT GGGCAGATCCCAGGGCAGCCCAATGGATCCAATGGTGATGAAGAGACCTCAGTTGTATGGCATGGGCAGTAACCCTCATTCTCAGCCTCA GCAGAGCAGTCCGTACCCAGGAGGTTCCTATGGCCCTCCAGGCCCACAGCGGTATCCAATTGGCATCCAGGGTCGGACTCCCGGGGCCAT GGCCGGAATGCAGTACCCTCAGCAGCAGATGCCACCTCAGTATGGACAGCAAGGTGTGAGTGGTTACTGCCAGCAGGGCCAACAGCCATA TTACAGCCAGCAGCCGCAGCCCCCGCACCTCCCACCCCAGGCGCAGTATCTGCCGTCCCAGTCCCAGCAGAGGTACCAGCCGCAGCAGGA CATGTCTCAGGAAGGCTATGGAACTAGATCTCAACCTCCTCTGGCCCCCGGAAAACCTAACCATGAAGACTTGAACTTAATACAGCAAGA AAGACCATCAAGTTTACCAGAAACAGACCTTATCCTGACTTTTAATATATCAATGCACCGATCTTGGTGGATGGAGAATGGACCAGGATG TACGGTGACGTCAGTGACACCTGCCCCAGACTGGGCCCCAGAAGACCATCGCTACATCACGGTCTCAGGGTGTTTGCTGGAGTACCAGTA CATAGAAGTGGCTCATAGTTCCCTCCAGATTGTCCTCGCACTGGCAGGTTTCATCTACGCCTGTTATGTTGTGAAATGTATAACTGAAGA AGAGGACAGCTTTGATTTCATAGGTGGCTTTGACTCTTATGGCTATCAAGGGCCTCAGAAGACATCTCATTTACAACTACAGCCTATGTA CATGTCAAAATAATACAGATGACTTCAGTATGTCAGCCCATGGACCTTTCAAAGAACTTTTTTCGCAGTGGCCTCCTGCATTTCATGAAG AGCAAGAAGCAACTGAGTTTAAATACATACACGTATTAACAAAACAAATGCAAAGCCTCTACATACAACACTGACACACACACACACACA CACGTGAGCACGCACACACCAATTCCACTTGACCTCCTCTTTCTAACTGAAACAGACAAATATGCAGGACACGCCCATCTTGGATTTCCT GAAAGCAGGCCCCTTTCTCCCAGGCCTTCGGAAAGTTCAGAAGGAGATGTGTTGATGCCCAACGGTTGCCGGCCATTGCTAACTCCTCTG CAGCCCAGCGGGTTGGCCTCTGTGAGCTGGGAAGTCATCCAAGGCACATTAGTTTGAGAGCTCTGTCTTCTGCACTCCATACATCTTGAC AGCACCACGGCTACTTAGGCAATGTAATTGCAAAAACAAACGAAACATGCCATGATGACCTTGTACCCGAAGTCCGAAATGGCAGATTGT TTGCCCTAAAGGCTGTACCGTACATACTTGCCTTAACCCACCTACATTGTGTGCCCAAGAATCCCAGTGCAGACAGCCCCAAGCCACTTC CTTTAGGCTAACACACTGCTATTAATTCTAAATGAGTTCAAGCCTGTGTGTTTCATGTCAAGGAACAAGACTGAATAGCTCAGCAGCTCC AGTGTGTCTGTATAAGAAAAGTGCCAGCTGTCAGCAAAGTGCTTTACCATAGAGCCTCCATGGTGGCCCAAAAGAGACAAGTAACAATAA CAATATCTGTCATCTTTATAATTCTAAAATATGGTCTTCTGATGGGGTTGGGGGTGGGACAGAGTAAAACATTAATCTACTAAACTCACT AATCTACTGGAAGCAGAAAGCAGAAATGGCTTAACTCGCTGATTTTATTTATTGCACTCTAGGTGCTTTTATAAGTATCTACAATTGTCT TTTACTAACACAGAAACAGCTGTGGATTTCTATCAACAGAGGCCAACTGAATATGTATTAGCTATGTTTACTCTTTTATATCTAATTCAA ATTGAAGACTCTACGTAGGTGAACTTTTTTTTCTAACTTCAGTGTACAAGATTATTTTTGGGTGGGTGGGGGGAGCAATAGTTACCTTGG TGAAATATTTGAGATCTTTTTGTGGTATTTGGGGGATCTGTTGTGTGTTAGAGTGTATTTCAACCCTCCTGTATTATTGTGGCAGAAAAA CTGTGCTGTTAACGTAGTTGGCAACAAGATGCCTTTGGAAACTTAATAGTAGGTCAATGTGTTTATGGAATATTCTGAACTCCTAGGAAA TTTTGTTCTATATTTTAAAGCATTATTTGGTTTTCATTAGTGGAAAACATAATTAGCTCAGGCTTATCAGGAACTCAGATGAGTCATATT TTTTCTTGGATACACCTGAGACATTAAACTCAATTTTTTAAAACTAACACACCATTCTTTAGTGCAGTGAAATTCAAAAGTTATAAATTG GCATGGAGCAAGTTTTTTCTTTGATTTCTATGAAGTTTCAGTTATTGGATTAAACTACTTTAGACCTTAGCCCATTCGGATCTACATACA CCAGTTCCATTACTGTGTTTGCATGGATGTCTGTAATATACACACACATATACATTTGAAAATGATAGAGGAACAAAGAGTTATCATTGG AACATAAACTATAAAAAAAGATTCCAGCACTCATGCTTGAATTTCCTCAGTGGGTTGTATGTTTGTCCATACATACATGTATAAAATAAA CAATTGCAAACATGTCAGTGAAACCTCATATGACGCTTTTATAGTGACATGCATGTTTAATGTATGAGAAAAATATTTACCAATGTATTG TCCTTTGAGTCCTAAAATATAAATACTATACATGCCAGAAACTATTCTAATGTGACTTTTAATGTGACTATTAAAAATGAGGTTTCACTG TACATTCATGCTGTGATGCGATTTCACCTTATAGTACATGTATATGGGTACAGAAAATAAACCCTCATATTCCTGCTTTAAGTTACAGCC AAGTTTTCACCAAATCACAACTATCATCACCACCAAAATTTTAAAAAACAGTAGTTGACTACTAAAAAGTAATCAGTGGACAGGTCATTT ATGGGATCAGTAAGCACAGTAGTGTGATTATATCTGGGAAAACATCTACAGTTGTACAGCTGTATCCTCCTCAAAATCCGGTGAAGAATG CTATTTTTCTAGGCTGAGCTTTTGTTATAAACTTAATATTCAGAAGGCAAGGGTTATGATCCTGATGTGTCCTTTTTTTTTGCATTTGTT AATTCTGAATGTATTTTTAACAGAATGATTTTTTTGACTTACTAGTGTAATTTGCATTTTAAAAATAAATGGAAAAATACTTAAGTTTCT GAGCCATCCCT >6409_6409_3_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000350026_NKAIN2_chr6_124979332_ENST00000368417_length(amino acids)=783AA_BP=664 MAHNAGAAAAAGTHSAKSGGSEAALKEGGSAAALSSSSSSSAAAAAASSSSSSGPGSAMETGLLPNHKLKTVGEAPAAPPHQQHHHHHHA HHHHHHAHHLHHHHALQQQLNQFQQQQQQQQQQQQQQQQQQHPISNNNSLGGAGGGAPQPGPDMEQPQHGGAKDSAAGGQADPPGPPLLS KPGDEDDAPPKMGEPAGGRYEHPGLGALGTQQPPVAVPGGGGGPAAVPEFNNYYGSAAPASGGPGGRAGPCFDQHGGQQSPGMGMMHSAS AAAAGAPGSMDPLQNSHEGYPNSQCNHYPGYSRPGAGGGGGGGGGGGGGSGGGGGGGGAGAGGAGAGAVAAAAAAAAAAAGGGGGGGYGG SSAGYGVLSSPRQQGGGMMMGPGGGGAASLSKAAAGSAAGGFQRFAGQNQHPSGATPTLNQLLTSPSPMMRSYGGSYPEYSSPSAPPPPP SQPQSQAAAAGAAAGGQQAAAGMGLGKDMGAQYAAASPAWAAAQQRSHPAMSPGTPGPTMGRSQGSPMDPMVMKRPQLYGMGSNPHSQPQ QSSPYPGGSYGPPGPQRYPIGIQGRTPGAMAGMQYPQQQMPPQYGQQGVSGYCQQGQQPYYSQQPQPPHLPPQAQYLPSQSQQRYQPQQD MSQEGYGTRSQPPLAPGKPNHEDLNLIQQERPSSLPETDLILTFNISMHRSWWMENGPGCTVTSVTPAPDWAPEDHRYITVSGCLLEYQY IEVAHSSLQIVLALAGFIYACYVVKCITEEEDSFDFIGGFDSYGYQGPQKTSHLQLQPMYMSK -------------------------------------------------------------- >6409_6409_4_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000367148_NKAIN2_chr6_124979332_ENST00000368417_length(transcript)=4780nt_BP=1998nt ATGGCCCATAACGCGGGCGCCGCGGCCGCCGCCGGCACCCACAGCGCCAAGAGCGGCGGCTCCGAGGCGGCTCTCAAGGAGGGTGGAAGC GCCGCCGCGCTGTCCTCCTCCTCCTCCTCCTCCGCGGCGGCAGCGGCGGCATCCTCTTCCTCCTCGTCGGGCCCGGGCTCGGCCATGGAG ACGGGGCTGCTCCCCAACCACAAACTGAAAACCGTTGGCGAAGCCCCCGCCGCGCCGCCCCACCAGCAGCACCACCACCACCACCATGCC CACCACCACCACCACCATGCCCACCACCTCCACCACCACCACGCACTACAGCAGCAGCTAAACCAGTTCCAGCAGCAGCAGCAGCAGCAG CAACAGCAGCAGCAGCAGCAGCAGCAACAGCAACATCCCATTTCCAACAACAACAGCTTGGGCGGCGCGGGCGGCGGCGCGCCTCAGCCC GGCCCCGACATGGAGCAGCCGCAACATGGAGGCGCCAAGGACAGTGCTGCGGGCGGCCAGGCCGACCCCCCGGGCCCGCCGCTGCTGAGC AAGCCGGGCGACGAGGACGACGCGCCGCCCAAGATGGGGGAGCCGGCGGGCGGCCGCTACGAGCACCCGGGCTTGGGCGCCCTGGGCACG CAGCAGCCGCCGGTCGCCGTGCCCGGGGGCGGCGGCGGCCCGGCGGCCGTCCCGGAGTTTAATAATTACTATGGCAGCGCTGCCCCTGCG AGCGGCGGCCCCGGCGGCCGCGCTGGGCCTTGCTTTGATCAACATGGCGGACAACAAAGCCCCGGGATGGGGATGATGCACTCCGCCTCC GCCGCCGCCGCCGGGGCCCCCGGCAGCATGGACCCCCTGCAGAACTCCCACGAAGGGTACCCCAACAGCCAGTGCAACCATTATCCGGGC TACAGCCGGCCCGGCGCGGGCGGCGGCGGCGGCGGCGGCGGCGGAGGAGGAGGAGGCAGCGGAGGAGGAGGAGGAGGAGGAGGAGCAGGA GCAGGAGGAGCAGGAGCGGGAGCTGTGGCGGCGGCGGCCGCGGCGGCGGCGGCAGCAGCAGGAGGCGGCGGCGGCGGCGGCTATGGGGGC TCGTCCGCGGGGTACGGGGTGCTGAGCTCCCCCCGGCAGCAGGGCGGCGGCATGATGATGGGCCCCGGGGGCGGCGGGGCCGCGAGCCTC AGCAAGGCGGCCGCCGGCTCGGCGGCGGGGGGCTTCCAGCGCTTCGCCGGCCAGAACCAGCACCCGTCGGGGGCCACCCCGACCCTCAAT CAGCTGCTCACCTCGCCCAGCCCCATGATGCGGAGCTACGGCGGCAGCTACCCCGAGTACAGCAGCCCCAGCGCGCCGCCGCCGCCGCCG TCGCAGCCCCAGTCCCAGGCGGCGGCGGCGGGGGCGGCGGCGGGCGGCCAGCAGGCGGCCGCGGGCATGGGCTTGGGCAAGGACATGGGC GCCCAGTACGCCGCTGCCAGCCCGGCCTGGGCGGCCGCGCAACAAAGGAGTCACCCGGCGATGAGCCCCGGCACCCCCGGACCGACCATG GGCAGATCCCAGGGCAGCCCAATGGATCCAATGGTGATGAAGAGACCTCAGTTGTATGGCATGGGCAGTAACCCTCATTCTCAGCCTCAG CAGAGCAGTCCGTACCCAGGAGGTTCCTATGGCCCTCCAGGCCCACAGCGGTATCCAATTGGCATCCAGGGTCGGACTCCCGGGGCCATG GCCGGAATGCAGTACCCTCAGCAGCAGATGCCACCTCAGTATGGACAGCAAGGTGTGAGTGGTTACTGCCAGCAGGGCCAACAGCCATAT TACAGCCAGCAGCCGCAGCCCCCGCACCTCCCACCCCAGGCGCAGTATCTGCCGTCCCAGTCCCAGCAGAGGTACCAGCCGCAGCAGGAC ATGTCTCAGGAAGGCTATGGAACTAGATCTCAACCTCCTCTGGCCCCCGGAAAACCTAACCATGAAGACTTGAACTTAATACAGCAAGAA AGACCATCAAGTTTACCAGAAACAGACCTTATCCTGACTTTTAATATATCAATGCACCGATCTTGGTGGATGGAGAATGGACCAGGATGT ACGGTGACGTCAGTGACACCTGCCCCAGACTGGGCCCCAGAAGACCATCGCTACATCACGGTCTCAGGGTGTTTGCTGGAGTACCAGTAC ATAGAAGTGGCTCATAGTTCCCTCCAGATTGTCCTCGCACTGGCAGGTTTCATCTACGCCTGTTATGTTGTGAAATGTATAACTGAAGAA GAGGACAGCTTTGATTTCATAGGTGGCTTTGACTCTTATGGCTATCAAGGGCCTCAGAAGACATCTCATTTACAACTACAGCCTATGTAC ATGTCAAAATAATACAGATGACTTCAGTATGTCAGCCCATGGACCTTTCAAAGAACTTTTTTCGCAGTGGCCTCCTGCATTTCATGAAGA GCAAGAAGCAACTGAGTTTAAATACATACACGTATTAACAAAACAAATGCAAAGCCTCTACATACAACACTGACACACACACACACACAC ACGTGAGCACGCACACACCAATTCCACTTGACCTCCTCTTTCTAACTGAAACAGACAAATATGCAGGACACGCCCATCTTGGATTTCCTG AAAGCAGGCCCCTTTCTCCCAGGCCTTCGGAAAGTTCAGAAGGAGATGTGTTGATGCCCAACGGTTGCCGGCCATTGCTAACTCCTCTGC AGCCCAGCGGGTTGGCCTCTGTGAGCTGGGAAGTCATCCAAGGCACATTAGTTTGAGAGCTCTGTCTTCTGCACTCCATACATCTTGACA GCACCACGGCTACTTAGGCAATGTAATTGCAAAAACAAACGAAACATGCCATGATGACCTTGTACCCGAAGTCCGAAATGGCAGATTGTT TGCCCTAAAGGCTGTACCGTACATACTTGCCTTAACCCACCTACATTGTGTGCCCAAGAATCCCAGTGCAGACAGCCCCAAGCCACTTCC TTTAGGCTAACACACTGCTATTAATTCTAAATGAGTTCAAGCCTGTGTGTTTCATGTCAAGGAACAAGACTGAATAGCTCAGCAGCTCCA GTGTGTCTGTATAAGAAAAGTGCCAGCTGTCAGCAAAGTGCTTTACCATAGAGCCTCCATGGTGGCCCAAAAGAGACAAGTAACAATAAC AATATCTGTCATCTTTATAATTCTAAAATATGGTCTTCTGATGGGGTTGGGGGTGGGACAGAGTAAAACATTAATCTACTAAACTCACTA ATCTACTGGAAGCAGAAAGCAGAAATGGCTTAACTCGCTGATTTTATTTATTGCACTCTAGGTGCTTTTATAAGTATCTACAATTGTCTT TTACTAACACAGAAACAGCTGTGGATTTCTATCAACAGAGGCCAACTGAATATGTATTAGCTATGTTTACTCTTTTATATCTAATTCAAA TTGAAGACTCTACGTAGGTGAACTTTTTTTTCTAACTTCAGTGTACAAGATTATTTTTGGGTGGGTGGGGGGAGCAATAGTTACCTTGGT GAAATATTTGAGATCTTTTTGTGGTATTTGGGGGATCTGTTGTGTGTTAGAGTGTATTTCAACCCTCCTGTATTATTGTGGCAGAAAAAC TGTGCTGTTAACGTAGTTGGCAACAAGATGCCTTTGGAAACTTAATAGTAGGTCAATGTGTTTATGGAATATTCTGAACTCCTAGGAAAT TTTGTTCTATATTTTAAAGCATTATTTGGTTTTCATTAGTGGAAAACATAATTAGCTCAGGCTTATCAGGAACTCAGATGAGTCATATTT TTTCTTGGATACACCTGAGACATTAAACTCAATTTTTTAAAACTAACACACCATTCTTTAGTGCAGTGAAATTCAAAAGTTATAAATTGG CATGGAGCAAGTTTTTTCTTTGATTTCTATGAAGTTTCAGTTATTGGATTAAACTACTTTAGACCTTAGCCCATTCGGATCTACATACAC CAGTTCCATTACTGTGTTTGCATGGATGTCTGTAATATACACACACATATACATTTGAAAATGATAGAGGAACAAAGAGTTATCATTGGA ACATAAACTATAAAAAAAGATTCCAGCACTCATGCTTGAATTTCCTCAGTGGGTTGTATGTTTGTCCATACATACATGTATAAAATAAAC AATTGCAAACATGTCAGTGAAACCTCATATGACGCTTTTATAGTGACATGCATGTTTAATGTATGAGAAAAATATTTACCAATGTATTGT CCTTTGAGTCCTAAAATATAAATACTATACATGCCAGAAACTATTCTAATGTGACTTTTAATGTGACTATTAAAAATGAGGTTTCACTGT ACATTCATGCTGTGATGCGATTTCACCTTATAGTACATGTATATGGGTACAGAAAATAAACCCTCATATTCCTGCTTTAAGTTACAGCCA AGTTTTCACCAAATCACAACTATCATCACCACCAAAATTTTAAAAAACAGTAGTTGACTACTAAAAAGTAATCAGTGGACAGGTCATTTA TGGGATCAGTAAGCACAGTAGTGTGATTATATCTGGGAAAACATCTACAGTTGTACAGCTGTATCCTCCTCAAAATCCGGTGAAGAATGC TATTTTTCTAGGCTGAGCTTTTGTTATAAACTTAATATTCAGAAGGCAAGGGTTATGATCCTGATGTGTCCTTTTTTTTTGCATTTGTTA ATTCTGAATGTATTTTTAACAGAATGATTTTTTTGACTTACTAGTGTAATTTGCATTTTAAAAATAAATGGAAAAATACTTAAGTTTCTG AGCCATCCCT >6409_6409_4_ARID1B-NKAIN2_ARID1B_chr6_157256710_ENST00000367148_NKAIN2_chr6_124979332_ENST00000368417_length(amino acids)=783AA_BP=664 MAHNAGAAAAAGTHSAKSGGSEAALKEGGSAAALSSSSSSSAAAAAASSSSSSGPGSAMETGLLPNHKLKTVGEAPAAPPHQQHHHHHHA HHHHHHAHHLHHHHALQQQLNQFQQQQQQQQQQQQQQQQQQHPISNNNSLGGAGGGAPQPGPDMEQPQHGGAKDSAAGGQADPPGPPLLS KPGDEDDAPPKMGEPAGGRYEHPGLGALGTQQPPVAVPGGGGGPAAVPEFNNYYGSAAPASGGPGGRAGPCFDQHGGQQSPGMGMMHSAS AAAAGAPGSMDPLQNSHEGYPNSQCNHYPGYSRPGAGGGGGGGGGGGGGSGGGGGGGGAGAGGAGAGAVAAAAAAAAAAAGGGGGGGYGG SSAGYGVLSSPRQQGGGMMMGPGGGGAASLSKAAAGSAAGGFQRFAGQNQHPSGATPTLNQLLTSPSPMMRSYGGSYPEYSSPSAPPPPP SQPQSQAAAAGAAAGGQQAAAGMGLGKDMGAQYAAASPAWAAAQQRSHPAMSPGTPGPTMGRSQGSPMDPMVMKRPQLYGMGSNPHSQPQ QSSPYPGGSYGPPGPQRYPIGIQGRTPGAMAGMQYPQQQMPPQYGQQGVSGYCQQGQQPYYSQQPQPPHLPPQAQYLPSQSQQRYQPQQD MSQEGYGTRSQPPLAPGKPNHEDLNLIQQERPSSLPETDLILTFNISMHRSWWMENGPGCTVTSVTPAPDWAPEDHRYITVSGCLLEYQY IEVAHSSLQIVLALAGFIYACYVVKCITEEEDSFDFIGGFDSYGYQGPQKTSHLQLQPMYMSK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ARID1B-NKAIN2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ARID1B-NKAIN2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ARID1B-NKAIN2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ARID1B | C0265338 | Coffin-Siris syndrome | 7 | CLINGEN;CTD_human;GENOMICS_ENGLAND |
| Hgene | ARID1B | C1535926 | Neurodevelopmental Disorders | 2 | CTD_human |
| Hgene | ARID1B | C3281201 | MENTAL RETARDATION, AUTOSOMAL DOMINANT 12 | 2 | GENOMICS_ENGLAND |
| Hgene | ARID1B | C0014544 | Epilepsy | 1 | CTD_human |
| Hgene | ARID1B | C0019569 | Hirschsprung Disease | 1 | GENOMICS_ENGLAND |
| Hgene | ARID1B | C0027819 | Neuroblastoma | 1 | CTD_human |
| Hgene | ARID1B | C0086237 | Epilepsy, Cryptogenic | 1 | CTD_human |
| Hgene | ARID1B | C0236018 | Aura | 1 | CTD_human |
| Hgene | ARID1B | C0751111 | Awakening Epilepsy | 1 | CTD_human |
| Hgene | ARID1B | C2239176 | Liver carcinoma | 1 | CGI;CTD_human |
| Hgene | ARID1B | C3150215 | CHROMOSOME 6q24-q25 DELETION SYNDROME | 1 | ORPHANET |
| Tgene | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
