
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:BCAT2-AK1 (FusionGDB2 ID:HG587TG203) |
Fusion Gene Summary for BCAT2-AK1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: BCAT2-AK1 | Fusion gene ID: hg587tg203 | Hgene | Tgene | Gene symbol | BCAT2 | AK1 | Gene ID | 587 | 203 |
| Gene name | branched chain amino acid transaminase 2 | adenylate kinase 1 | |
| Synonyms | BCAM|BCATM|BCT2|PP18 | HTL-S-58j | |
| Cytomap | ('BCAT2')('AK1') 19q13.33 | 9q34.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | branched-chain-amino-acid aminotransferase, mitochondrialbranched chain amino-acid transaminase 2, mitochondrialbranched chain aminotransferase 2, mitochondrialplacental protein 18 | adenylate kinase isoenzyme 1ATP-AMP transphosphorylase 1ATP:AMP phosphotransferaseadenylate monophosphate kinaseepididymis secretory sperm binding proteinmyokinasetestis secretory sperm binding protein Li 58j | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | P00568 | |
| Ensembl transtripts involved in fusion gene | ENST00000402551, ENST00000601496, ENST00000316273, ENST00000545387, ENST00000598162, ENST00000599246, ENST00000597011, | ||
| Fusion gene scores | * DoF score | 10 X 9 X 9=810 | 3 X 2 X 3=18 |
| # samples | 11 | 3 | |
| ** MAII score | log2(11/810*10)=-2.88041838424733 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: BCAT2 [Title/Abstract] AND AK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | BCAT2(49314240)-AK1(130630791), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | BCAT2-AK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BCAT2-AK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BCAT2-AK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BCAT2-AK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | AK1 | GO:0006165 | nucleoside diphosphate phosphorylation | 23416111 |
| Tgene | AK1 | GO:0009142 | nucleoside triphosphate biosynthetic process | 23416111 |
 Fusion gene breakpoints across BCAT2 (5'-gene) Fusion gene breakpoints across BCAT2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
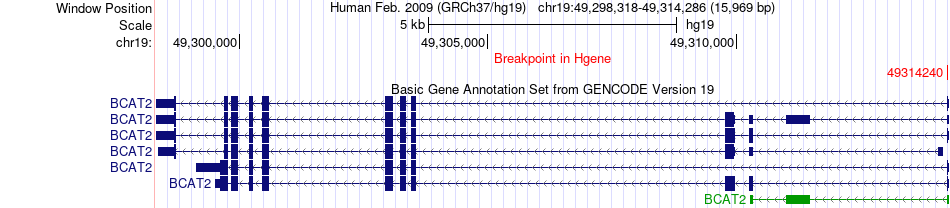 |
 Fusion gene breakpoints across AK1 (3'-gene) Fusion gene breakpoints across AK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
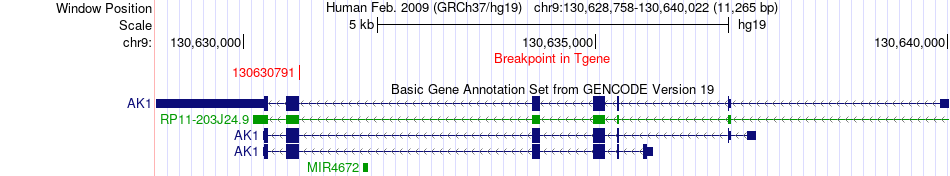 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-IM-A3ED | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
Top |
Fusion Gene ORF analysis for BCAT2-AK1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000402551 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| 5UTR-3CDS | ENST00000402551 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| 5UTR-3CDS | ENST00000402551 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| 5UTR-3CDS | ENST00000601496 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| 5UTR-3CDS | ENST00000601496 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| 5UTR-3CDS | ENST00000601496 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000316273 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000316273 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000316273 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000545387 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000545387 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000545387 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000598162 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000598162 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000598162 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000599246 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000599246 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| In-frame | ENST00000599246 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| intron-3CDS | ENST00000597011 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| intron-3CDS | ENST00000597011 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
| intron-3CDS | ENST00000597011 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000545387 | BCAT2 | chr19 | 49314240 | - | ENST00000373176 | AK1 | chr9 | 130630791 | - | 1818 | 29 | 960 | 559 | 133 |
| ENST00000545387 | BCAT2 | chr19 | 49314240 | - | ENST00000373156 | AK1 | chr9 | 130630791 | - | 299 | 29 | 5 | 289 | 94 |
| ENST00000545387 | BCAT2 | chr19 | 49314240 | - | ENST00000223836 | AK1 | chr9 | 130630791 | - | 297 | 29 | 5 | 289 | 94 |
| ENST00000316273 | BCAT2 | chr19 | 49314240 | - | ENST00000373176 | AK1 | chr9 | 130630791 | - | 1826 | 37 | 968 | 567 | 133 |
| ENST00000316273 | BCAT2 | chr19 | 49314240 | - | ENST00000373156 | AK1 | chr9 | 130630791 | - | 307 | 37 | 13 | 297 | 94 |
| ENST00000316273 | BCAT2 | chr19 | 49314240 | - | ENST00000223836 | AK1 | chr9 | 130630791 | - | 305 | 37 | 13 | 297 | 94 |
| ENST00000599246 | BCAT2 | chr19 | 49314240 | - | ENST00000373176 | AK1 | chr9 | 130630791 | - | 1835 | 46 | 977 | 576 | 133 |
| ENST00000599246 | BCAT2 | chr19 | 49314240 | - | ENST00000373156 | AK1 | chr9 | 130630791 | - | 316 | 46 | 22 | 306 | 94 |
| ENST00000599246 | BCAT2 | chr19 | 49314240 | - | ENST00000223836 | AK1 | chr9 | 130630791 | - | 314 | 46 | 22 | 306 | 94 |
| ENST00000598162 | BCAT2 | chr19 | 49314240 | - | ENST00000373176 | AK1 | chr9 | 130630791 | - | 1833 | 44 | 975 | 574 | 133 |
| ENST00000598162 | BCAT2 | chr19 | 49314240 | - | ENST00000373156 | AK1 | chr9 | 130630791 | - | 314 | 44 | 20 | 304 | 94 |
| ENST00000598162 | BCAT2 | chr19 | 49314240 | - | ENST00000223836 | AK1 | chr9 | 130630791 | - | 312 | 44 | 20 | 304 | 94 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000545387 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.9040109 | 0.09598913 |
| ENST00000545387 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.16643089 | 0.8335691 |
| ENST00000545387 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.16292875 | 0.8370713 |
| ENST00000316273 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.9285581 | 0.07144186 |
| ENST00000316273 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.12977558 | 0.87022436 |
| ENST00000316273 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.12804803 | 0.871952 |
| ENST00000599246 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.90919787 | 0.09080211 |
| ENST00000599246 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.1247803 | 0.8752197 |
| ENST00000599246 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.12257613 | 0.8774238 |
| ENST00000598162 | ENST00000373176 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.8892345 | 0.11076548 |
| ENST00000598162 | ENST00000373156 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.12780029 | 0.87219965 |
| ENST00000598162 | ENST00000223836 | BCAT2 | chr19 | 49314240 | - | AK1 | chr9 | 130630791 | - | 0.12539916 | 0.8746008 |
Top |
Fusion Genomic Features for BCAT2-AK1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
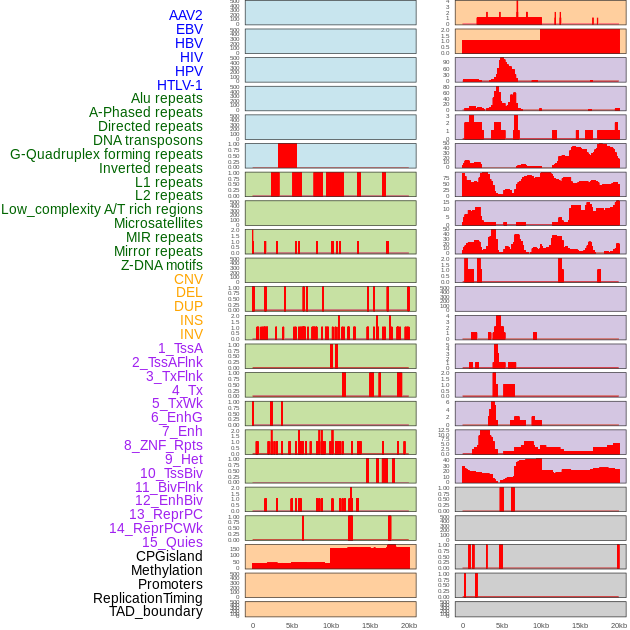 |
Top |
Fusion Protein Features for BCAT2-AK1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:49314240/chr9:130630791) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | AK1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Catalyzes the reversible transfer of the terminal phosphate group between ATP and AMP. Also displays broad nucleoside diphosphate kinase activity. Plays an important role in cellular energy homeostasis and in adenine nucleotide metabolism. {ECO:0000255|HAMAP-Rule:MF_03171, ECO:0000269|PubMed:23416111}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373156 | 4 | 7 | 131_141 | 108 | 195.0 | Region | LID | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373176 | 4 | 7 | 131_141 | 108 | 195.0 | Region | LID |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373156 | 4 | 7 | 18_23 | 108 | 195.0 | Nucleotide binding | ATP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373156 | 4 | 7 | 65_67 | 108 | 195.0 | Nucleotide binding | AMP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373156 | 4 | 7 | 94_97 | 108 | 195.0 | Nucleotide binding | AMP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373176 | 4 | 7 | 18_23 | 108 | 195.0 | Nucleotide binding | ATP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373176 | 4 | 7 | 65_67 | 108 | 195.0 | Nucleotide binding | AMP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373176 | 4 | 7 | 94_97 | 108 | 195.0 | Nucleotide binding | AMP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373156 | 4 | 7 | 38_67 | 108 | 195.0 | Region | NMP | |
| Tgene | AK1 | chr19:49314240 | chr9:130630791 | ENST00000373176 | 4 | 7 | 38_67 | 108 | 195.0 | Region | NMP |
Top |
Fusion Gene Sequence for BCAT2-AK1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >9282_9282_1_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000223836_length(transcript)=305nt_BP=37nt CGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGAC CCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGC CACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGT CTGCACCCACCTGGACGCCCTAAAGTAGCAACGCT >9282_9282_1_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000223836_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_2_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000373156_length(transcript)=307nt_BP=37nt CGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGAC CCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGC CACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGT CTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGG >9282_9282_2_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000373156_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_3_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000373176_length(transcript)=1826nt_BP=37nt CGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGAC CCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGC CACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGT CTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGGAGCCGCTTCCCCAGCTCAGAGCCCCGCCCCACCCCGTCCTGATTCGAGGTCCT CCTGGCCTGAGCGCAGCGCCTCCACCCTGCCCTGCTGAGCACAGACGGAGGAAGCCGCTTATCCTGTTTTCATGGACAGCTGAGCACTAA AGGAATTTCTAAGGACATTTGGTTTTACTGCTTTTTCTCTGCTTCCAGTTGGAGTTGATTCATGTGCTTGTGCCTACCTGGCCGCAAGTC CCCAGCCCCTCAACCCTCCGTTCCTCCTCAGCCTCCCTTTGCCAGCCACCCCTCCTCTAGCTCTGGTGGGAGGCCCGGGGCCCTTCCTCG CACAGGGCATGCCTGGCCTGAGGACCCGGCGCTGAGTGGCGGGGCCCCTGCTCCGAGGGGCTCATGTTCAGGCAGAACCGGTCCCAGCCT GGGCTCCTCTGCATCTTGCTCTGTGGCCTTGGCCCTGACCCCCATCGCTCTGAGCATATGTTCCATGCCTGGCCCTTGCCGGGGCCTGGA CTGCACAGGCAGCAAGGTCATGGTCTGAGTGGGGCTTCCTGGGCAGTTGGGGCGGCCCACGCCAGCTGGCCCAGTGGGTAGTGAATTGGC TTCCTTGACGCGAGAGGCTCTGAGGGTCTGAAAAGGGCATCTCAATGGCATGGGTGGGTGGGGAGTCAGTCATGTCACTGAAATTGAATG GGGGAGGCCCAATGAGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAAGCTGAGGCAGGAGGATCCCCTGAGGTCAGGAGTTCGAGAG CAGCCTGGCCAACATGGCAAAACCCCTTATTTACTAAAAATACAAAAATTAGCCGGGCATGGTGGCATGTGCCTGTATTCCCAGCTACTC ATGAGGCTGAGGCAGGAGAATGGCGTGAACCCGGGAGGTGGAGCTTGCAATGAGCCAAGATTGCGCCACTGCACTCCAGCCTGGGTGACA GAGCAAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAGAAATTGAATTGGGGGAGTCATGGTCAGGGGTGAGACCTGAAGGACCTCCCC CTGTGTGGCCCTGGACACAGCCCACCCTCTGTGAGCCGTTTCCAATTCTAAAACAGACTCAATGTCCCCCTCACCCCCACCTCAAGGTCA GGATGAGAACACACTGAGTGAGGAGTGGACGCTGTCCATTGCCACGGCCATGAGGGCTGGAGACCAGAACAGCATGGCCCGAAGCGTGCG GGGCCCCGGATGACTTGGGGACACCCCAGAATCCCCTGGGGAGAACCCTTCCTGCGCGCTTTCATTTTTTGACCTCATCACTGAGAAAGG CTCAATTTGGTGCTCACGTGTCCTTAACACCTGATCTGGCCCAAGCTGGGCTCCCTTTAAGCCAAGAGAGCCTCTTGTGGACCCCGCCTG CCCGAATGAAATCCGAACAGTTGGGGCTGTTCATGGCAAGTGGGGCTGGTTTTTCATTTCCATTGGTTATTTAAAGTTTCCTTTAAAATA AACGATTTTAAGTTATAAAAGGTGAA >9282_9282_3_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000316273_AK1_chr9_130630791_ENST00000373176_length(amino acids)=133AA_BP= MTPHPPMPLRCPFQTLRASRVKEANSLPTGPAGVGRPNCPGSPTQTMTLLPVQSRPRQGPGMEHMLRAMGVRAKATEQDAEEPRLGPVLP EHEPLGAGAPPLSAGSSGQACPVRGRAPGLPPELEEGWLAKGG -------------------------------------------------------------- >9282_9282_4_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000223836_length(transcript)=297nt_BP=29nt GGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGACCCAGCGGC TCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGCCACAGAAC CTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGTCTGCACCC ACCTGGACGCCCTAAAGTAGCAACGCT >9282_9282_4_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000223836_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_5_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000373156_length(transcript)=299nt_BP=29nt GGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGACCCAGCGGC TCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGCCACAGAAC CTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGTCTGCACCC ACCTGGACGCCCTAAAGTAGCAACGCTGG >9282_9282_5_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000373156_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_6_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000373176_length(transcript)=1818nt_BP=29nt GGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGACCATGACCCAGCGGC TCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATTACAAGGCCACAGAAC CTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCTCCCAGGTCTGCACCC ACCTGGACGCCCTAAAGTAGCAACGCTGGAGCCGCTTCCCCAGCTCAGAGCCCCGCCCCACCCCGTCCTGATTCGAGGTCCTCCTGGCCT GAGCGCAGCGCCTCCACCCTGCCCTGCTGAGCACAGACGGAGGAAGCCGCTTATCCTGTTTTCATGGACAGCTGAGCACTAAAGGAATTT CTAAGGACATTTGGTTTTACTGCTTTTTCTCTGCTTCCAGTTGGAGTTGATTCATGTGCTTGTGCCTACCTGGCCGCAAGTCCCCAGCCC CTCAACCCTCCGTTCCTCCTCAGCCTCCCTTTGCCAGCCACCCCTCCTCTAGCTCTGGTGGGAGGCCCGGGGCCCTTCCTCGCACAGGGC ATGCCTGGCCTGAGGACCCGGCGCTGAGTGGCGGGGCCCCTGCTCCGAGGGGCTCATGTTCAGGCAGAACCGGTCCCAGCCTGGGCTCCT CTGCATCTTGCTCTGTGGCCTTGGCCCTGACCCCCATCGCTCTGAGCATATGTTCCATGCCTGGCCCTTGCCGGGGCCTGGACTGCACAG GCAGCAAGGTCATGGTCTGAGTGGGGCTTCCTGGGCAGTTGGGGCGGCCCACGCCAGCTGGCCCAGTGGGTAGTGAATTGGCTTCCTTGA CGCGAGAGGCTCTGAGGGTCTGAAAAGGGCATCTCAATGGCATGGGTGGGTGGGGAGTCAGTCATGTCACTGAAATTGAATGGGGGAGGC CCAATGAGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAAGCTGAGGCAGGAGGATCCCCTGAGGTCAGGAGTTCGAGAGCAGCCTGG CCAACATGGCAAAACCCCTTATTTACTAAAAATACAAAAATTAGCCGGGCATGGTGGCATGTGCCTGTATTCCCAGCTACTCATGAGGCT GAGGCAGGAGAATGGCGTGAACCCGGGAGGTGGAGCTTGCAATGAGCCAAGATTGCGCCACTGCACTCCAGCCTGGGTGACAGAGCAAGA CTCCGTCTCAAAAAAAAAAAAAAAAAAAAAGAAATTGAATTGGGGGAGTCATGGTCAGGGGTGAGACCTGAAGGACCTCCCCCTGTGTGG CCCTGGACACAGCCCACCCTCTGTGAGCCGTTTCCAATTCTAAAACAGACTCAATGTCCCCCTCACCCCCACCTCAAGGTCAGGATGAGA ACACACTGAGTGAGGAGTGGACGCTGTCCATTGCCACGGCCATGAGGGCTGGAGACCAGAACAGCATGGCCCGAAGCGTGCGGGGCCCCG GATGACTTGGGGACACCCCAGAATCCCCTGGGGAGAACCCTTCCTGCGCGCTTTCATTTTTTGACCTCATCACTGAGAAAGGCTCAATTT GGTGCTCACGTGTCCTTAACACCTGATCTGGCCCAAGCTGGGCTCCCTTTAAGCCAAGAGAGCCTCTTGTGGACCCCGCCTGCCCGAATG AAATCCGAACAGTTGGGGCTGTTCATGGCAAGTGGGGCTGGTTTTTCATTTCCATTGGTTATTTAAAGTTTCCTTTAAAATAAACGATTT TAAGTTATAAAAGGTGAA >9282_9282_6_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000545387_AK1_chr9_130630791_ENST00000373176_length(amino acids)=133AA_BP= MTPHPPMPLRCPFQTLRASRVKEANSLPTGPAGVGRPNCPGSPTQTMTLLPVQSRPRQGPGMEHMLRAMGVRAKATEQDAEEPRLGPVLP EHEPLGAGAPPLSAGSSGQACPVRGRAPGLPPELEEGWLAKGG -------------------------------------------------------------- >9282_9282_7_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000223836_length(transcript)=312nt_BP=44nt AGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGA CCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATT ACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCT CCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCT >9282_9282_7_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000223836_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_8_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000373156_length(transcript)=314nt_BP=44nt AGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGA CCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATT ACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCT CCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGG >9282_9282_8_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000373156_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_9_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000373176_length(transcript)=1833nt_BP=44nt AGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGAGA CCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTATT ACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTTCT CCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGGAGCCGCTTCCCCAGCTCAGAGCCCCGCCCCACCCCGTCCTGATTCG AGGTCCTCCTGGCCTGAGCGCAGCGCCTCCACCCTGCCCTGCTGAGCACAGACGGAGGAAGCCGCTTATCCTGTTTTCATGGACAGCTGA GCACTAAAGGAATTTCTAAGGACATTTGGTTTTACTGCTTTTTCTCTGCTTCCAGTTGGAGTTGATTCATGTGCTTGTGCCTACCTGGCC GCAAGTCCCCAGCCCCTCAACCCTCCGTTCCTCCTCAGCCTCCCTTTGCCAGCCACCCCTCCTCTAGCTCTGGTGGGAGGCCCGGGGCCC TTCCTCGCACAGGGCATGCCTGGCCTGAGGACCCGGCGCTGAGTGGCGGGGCCCCTGCTCCGAGGGGCTCATGTTCAGGCAGAACCGGTC CCAGCCTGGGCTCCTCTGCATCTTGCTCTGTGGCCTTGGCCCTGACCCCCATCGCTCTGAGCATATGTTCCATGCCTGGCCCTTGCCGGG GCCTGGACTGCACAGGCAGCAAGGTCATGGTCTGAGTGGGGCTTCCTGGGCAGTTGGGGCGGCCCACGCCAGCTGGCCCAGTGGGTAGTG AATTGGCTTCCTTGACGCGAGAGGCTCTGAGGGTCTGAAAAGGGCATCTCAATGGCATGGGTGGGTGGGGAGTCAGTCATGTCACTGAAA TTGAATGGGGGAGGCCCAATGAGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAAGCTGAGGCAGGAGGATCCCCTGAGGTCAGGAGT TCGAGAGCAGCCTGGCCAACATGGCAAAACCCCTTATTTACTAAAAATACAAAAATTAGCCGGGCATGGTGGCATGTGCCTGTATTCCCA GCTACTCATGAGGCTGAGGCAGGAGAATGGCGTGAACCCGGGAGGTGGAGCTTGCAATGAGCCAAGATTGCGCCACTGCACTCCAGCCTG GGTGACAGAGCAAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAGAAATTGAATTGGGGGAGTCATGGTCAGGGGTGAGACCTGAAGGA CCTCCCCCTGTGTGGCCCTGGACACAGCCCACCCTCTGTGAGCCGTTTCCAATTCTAAAACAGACTCAATGTCCCCCTCACCCCCACCTC AAGGTCAGGATGAGAACACACTGAGTGAGGAGTGGACGCTGTCCATTGCCACGGCCATGAGGGCTGGAGACCAGAACAGCATGGCCCGAA GCGTGCGGGGCCCCGGATGACTTGGGGACACCCCAGAATCCCCTGGGGAGAACCCTTCCTGCGCGCTTTCATTTTTTGACCTCATCACTG AGAAAGGCTCAATTTGGTGCTCACGTGTCCTTAACACCTGATCTGGCCCAAGCTGGGCTCCCTTTAAGCCAAGAGAGCCTCTTGTGGACC CCGCCTGCCCGAATGAAATCCGAACAGTTGGGGCTGTTCATGGCAAGTGGGGCTGGTTTTTCATTTCCATTGGTTATTTAAAGTTTCCTT TAAAATAAACGATTTTAAGTTATAAAAGGTGAA >9282_9282_9_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000598162_AK1_chr9_130630791_ENST00000373176_length(amino acids)=133AA_BP= MTPHPPMPLRCPFQTLRASRVKEANSLPTGPAGVGRPNCPGSPTQTMTLLPVQSRPRQGPGMEHMLRAMGVRAKATEQDAEEPRLGPVLP EHEPLGAGAPPLSAGSSGQACPVRGRAPGLPPELEEGWLAKGG -------------------------------------------------------------- >9282_9282_10_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000223836_length(transcript)=314nt_BP=46nt ACAGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGA GACCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTA TTACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTT CTCCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCT >9282_9282_10_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000223836_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_11_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000373156_length(transcript)=316nt_BP=46nt ACAGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGA GACCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTA TTACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTT CTCCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGG >9282_9282_11_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000373156_length(amino acids)=94AA_BP=7 MAAAALGQIGQPTLLLYVDAGPETMTQRLLKRGETSGRVDDNEETIKKRLETYYKATEPVIAFYEKRGIVRKVNAEGSVDSVFSQVCTHL DALK -------------------------------------------------------------- >9282_9282_12_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000373176_length(transcript)=1835nt_BP=46nt ACAGTTACGCGCCGCACGGATCATGGCCGCAGCCGCTCTGGGGCAGATTGGACAGCCCACACTGCTGCTGTATGTGGACGCAGGCCCTGA GACCATGACCCAGCGGCTCTTGAAACGTGGAGAGACCAGCGGGCGTGTGGACGACAATGAGGAGACCATCAAAAAGCGGCTGGAGACCTA TTACAAGGCCACAGAACCTGTCATCGCCTTCTATGAGAAACGTGGCATTGTGCGCAAGGTCAACGCTGAGGGCTCCGTGGACAGTGTCTT CTCCCAGGTCTGCACCCACCTGGACGCCCTAAAGTAGCAACGCTGGAGCCGCTTCCCCAGCTCAGAGCCCCGCCCCACCCCGTCCTGATT CGAGGTCCTCCTGGCCTGAGCGCAGCGCCTCCACCCTGCCCTGCTGAGCACAGACGGAGGAAGCCGCTTATCCTGTTTTCATGGACAGCT GAGCACTAAAGGAATTTCTAAGGACATTTGGTTTTACTGCTTTTTCTCTGCTTCCAGTTGGAGTTGATTCATGTGCTTGTGCCTACCTGG CCGCAAGTCCCCAGCCCCTCAACCCTCCGTTCCTCCTCAGCCTCCCTTTGCCAGCCACCCCTCCTCTAGCTCTGGTGGGAGGCCCGGGGC CCTTCCTCGCACAGGGCATGCCTGGCCTGAGGACCCGGCGCTGAGTGGCGGGGCCCCTGCTCCGAGGGGCTCATGTTCAGGCAGAACCGG TCCCAGCCTGGGCTCCTCTGCATCTTGCTCTGTGGCCTTGGCCCTGACCCCCATCGCTCTGAGCATATGTTCCATGCCTGGCCCTTGCCG GGGCCTGGACTGCACAGGCAGCAAGGTCATGGTCTGAGTGGGGCTTCCTGGGCAGTTGGGGCGGCCCACGCCAGCTGGCCCAGTGGGTAG TGAATTGGCTTCCTTGACGCGAGAGGCTCTGAGGGTCTGAAAAGGGCATCTCAATGGCATGGGTGGGTGGGGAGTCAGTCATGTCACTGA AATTGAATGGGGGAGGCCCAATGAGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAAGCTGAGGCAGGAGGATCCCCTGAGGTCAGGA GTTCGAGAGCAGCCTGGCCAACATGGCAAAACCCCTTATTTACTAAAAATACAAAAATTAGCCGGGCATGGTGGCATGTGCCTGTATTCC CAGCTACTCATGAGGCTGAGGCAGGAGAATGGCGTGAACCCGGGAGGTGGAGCTTGCAATGAGCCAAGATTGCGCCACTGCACTCCAGCC TGGGTGACAGAGCAAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAGAAATTGAATTGGGGGAGTCATGGTCAGGGGTGAGACCTGAAG GACCTCCCCCTGTGTGGCCCTGGACACAGCCCACCCTCTGTGAGCCGTTTCCAATTCTAAAACAGACTCAATGTCCCCCTCACCCCCACC TCAAGGTCAGGATGAGAACACACTGAGTGAGGAGTGGACGCTGTCCATTGCCACGGCCATGAGGGCTGGAGACCAGAACAGCATGGCCCG AAGCGTGCGGGGCCCCGGATGACTTGGGGACACCCCAGAATCCCCTGGGGAGAACCCTTCCTGCGCGCTTTCATTTTTTGACCTCATCAC TGAGAAAGGCTCAATTTGGTGCTCACGTGTCCTTAACACCTGATCTGGCCCAAGCTGGGCTCCCTTTAAGCCAAGAGAGCCTCTTGTGGA CCCCGCCTGCCCGAATGAAATCCGAACAGTTGGGGCTGTTCATGGCAAGTGGGGCTGGTTTTTCATTTCCATTGGTTATTTAAAGTTTCC TTTAAAATAAACGATTTTAAGTTATAAAAGGTGAA >9282_9282_12_BCAT2-AK1_BCAT2_chr19_49314240_ENST00000599246_AK1_chr9_130630791_ENST00000373176_length(amino acids)=133AA_BP= MTPHPPMPLRCPFQTLRASRVKEANSLPTGPAGVGRPNCPGSPTQTMTLLPVQSRPRQGPGMEHMLRAMGVRAKATEQDAEEPRLGPVLP EHEPLGAGAPPLSAGSSGQACPVRGRAPGLPPELEEGWLAKGG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for BCAT2-AK1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for BCAT2-AK1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for BCAT2-AK1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | BCAT2 | C0024776 | Maple Syrup Urine Disease | 1 | CTD_human |
| Hgene | BCAT2 | C0032927 | Precancerous Conditions | 1 | CTD_human |
| Hgene | BCAT2 | C0268568 | Classic Maple Syrup Urine Disease | 1 | CTD_human |
| Hgene | BCAT2 | C0268569 | Intermittent Maple Syrup Urine Disease | 1 | CTD_human |
| Hgene | BCAT2 | C0282313 | Condition, Preneoplastic | 1 | CTD_human |
| Hgene | BCAT2 | C0751285 | Maple Syrup Urine Disease, Thiamine Responsive | 1 | CTD_human |
| Hgene | BCAT2 | C1621920 | Intermediate Maple Syrup Urine Disease | 1 | CTD_human |
| Tgene | C2675459 | Adenylate Kinase Deficiency, Hemolytic Anemia Due To | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0151744 | Myocardial Ischemia | 1 | CTD_human | |
| Tgene | C0334634 | Malignant lymphoma, lymphocytic, intermediate differentiation, diffuse | 1 | CTD_human | |
| Tgene | C0751958 | Lymphoma, Lymphocytic, Intermediate | 1 | CTD_human |
