
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACTB-COL17A1 (FusionGDB2 ID:HG60TG1308) |
Fusion Gene Summary for ACTB-COL17A1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACTB-COL17A1 | Fusion gene ID: hg60tg1308 | Hgene | Tgene | Gene symbol | ACTB | COL17A1 | Gene ID | 60 | 1308 |
| Gene name | actin beta | collagen type XVII alpha 1 chain | |
| Synonyms | BRWS1|PS1TP5BP1 | BA16H23.2|BP180|BPA-2|BPAG2|ERED|LAD-1 | |
| Cytomap | ('ACTB')('COL17A1') 7p22.1 | 10q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | actin, cytoplasmic 1I(2)-actinPS1TP5-binding protein 1beta cytoskeletal actin | collagen alpha-1(XVII) chain180 kDa bullous pemphigoid antigen 2alpha 1 type XVII collagenbA16H23.2 (collagen, type XVII, alpha 1 (BP180))bullous pemphigoid antigen 2 (180kD)collagen XVII, alpha-1 polypeptidecollagen, type XVII, alpha 1type XVII co | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P60709 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000331789, ENST00000464611, | ||
| Fusion gene scores | * DoF score | 68 X 54 X 18=66096 | 4 X 5 X 5=100 |
| # samples | 83 | 6 | |
| ** MAII score | log2(83/66096*10)=-6.31530781862183 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/100*10)=-0.736965594166206 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ACTB [Title/Abstract] AND COL17A1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACTB(5569166)-COL17A1(105801102), # samples:1 ACTB(5567255)-COL17A1(105845629), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ACTB-COL17A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACTB-COL17A1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACTB-COL17A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ACTB-COL17A1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACTB | GO:0098974 | postsynaptic actin cytoskeleton organization | 18341992 |
| Tgene | COL17A1 | GO:0031581 | hemidesmosome assembly | 12482924 |
 Fusion gene breakpoints across ACTB (5'-gene) Fusion gene breakpoints across ACTB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
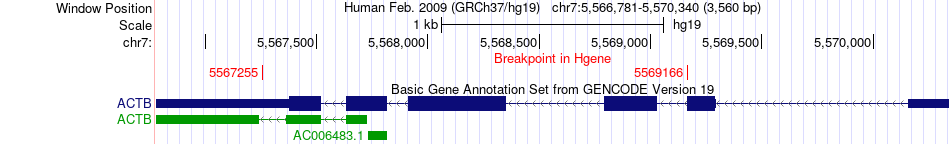 |
 Fusion gene breakpoints across COL17A1 (3'-gene) Fusion gene breakpoints across COL17A1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
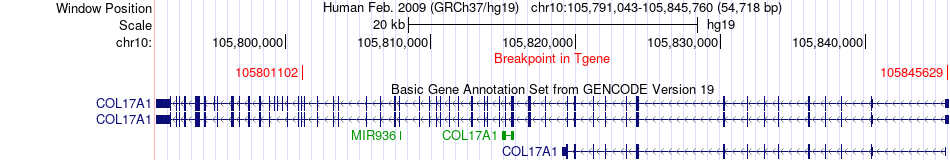 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CV-7104-01A | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| ChimerDB4 | LUAD | TCGA-78-7542-01A | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
Top |
Fusion Gene ORF analysis for ACTB-COL17A1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000331789 | ENST00000393211 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| 5CDS-intron | ENST00000331789 | ENST00000480127 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| In-frame | ENST00000331789 | ENST00000353479 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| In-frame | ENST00000331789 | ENST00000369733 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| intron-3CDS | ENST00000464611 | ENST00000353479 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| intron-3CDS | ENST00000464611 | ENST00000369733 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| intron-5UTR | ENST00000331789 | ENST00000353479 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-5UTR | ENST00000331789 | ENST00000369733 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-5UTR | ENST00000464611 | ENST00000353479 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-5UTR | ENST00000464611 | ENST00000369733 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-intron | ENST00000331789 | ENST00000393211 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-intron | ENST00000331789 | ENST00000480127 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-intron | ENST00000464611 | ENST00000393211 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| intron-intron | ENST00000464611 | ENST00000393211 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
| intron-intron | ENST00000464611 | ENST00000480127 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - |
| intron-intron | ENST00000464611 | ENST00000480127 | ACTB | chr7 | 5567255 | - | COL17A1 | chr10 | 105845629 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000331789 | ACTB | chr7 | 5569166 | - | ENST00000369733 | COL17A1 | chr10 | 105801102 | - | 2907 | 315 | 365 | 1957 | 530 |
| ENST00000331789 | ACTB | chr7 | 5569166 | - | ENST00000353479 | COL17A1 | chr10 | 105801102 | - | 3153 | 315 | 365 | 2203 | 612 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000331789 | ENST00000369733 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - | 0.0068612 | 0.99313873 |
| ENST00000331789 | ENST00000353479 | ACTB | chr7 | 5569166 | - | COL17A1 | chr10 | 105801102 | - | 0.005469699 | 0.99453026 |
Top |
Fusion Genomic Features for ACTB-COL17A1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
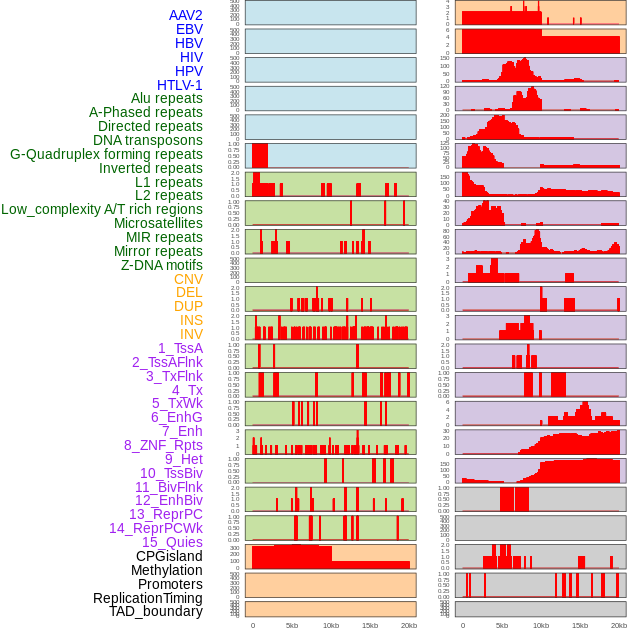 |
Top |
Fusion Protein Features for ACTB-COL17A1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:5569166/chr10:105801102) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACTB | . |
| FUNCTION: Actin is a highly conserved protein that polymerizes to produce filaments that form cross-linked networks in the cytoplasm of cells (PubMed:29581253). Actin exists in both monomeric (G-actin) and polymeric (F-actin) forms, both forms playing key functions, such as cell motility and contraction (PubMed:29581253). In addition to their role in the cytoplasmic cytoskeleton, G- and F-actin also localize in the nucleus, and regulate gene transcription and motility and repair of damaged DNA (PubMed:29925947). {ECO:0000269|PubMed:29581253, ECO:0000269|PubMed:29925947}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 1483_1497 | 868 | 1498.0 | Region | Note=Nonhelical region (NC1) | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 1483_1497 | 868 | 1416.0 | Region | Note=Nonhelical region (NC1) |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 1_566 | 868 | 1498.0 | Region | Note=Nonhelical region (NC16) | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 567_1482 | 868 | 1498.0 | Region | Note=Triple-helical region | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 1_566 | 868 | 1416.0 | Region | Note=Nonhelical region (NC16) | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 567_1482 | 868 | 1416.0 | Region | Note=Triple-helical region | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 1_467 | 868 | 1498.0 | Topological domain | Cytoplasmic | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 489_1497 | 868 | 1498.0 | Topological domain | Extracellular | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 1_467 | 868 | 1416.0 | Topological domain | Cytoplasmic | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 489_1497 | 868 | 1416.0 | Topological domain | Extracellular | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000353479 | 36 | 56 | 468_488 | 868 | 1498.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein | |
| Tgene | COL17A1 | chr7:5569166 | chr10:105801102 | ENST00000369733 | 36 | 52 | 468_488 | 868 | 1416.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for ACTB-COL17A1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1584_1584_1_ACTB-COL17A1_ACTB_chr7_5569166_ENST00000331789_COL17A1_chr10_105801102_ENST00000353479_length(transcript)=3153nt_BP=315nt GAGTGAGCGGCGCGGGGCCAATCAGCGTGCGCCGTTCCGAAAGTTGCCTTTTATGGCTCGAGCGGCCGCGGCGGCGCCCTATAAAACCCA GCGGCGCGACGCGCCACCACCGCCGAGACCGCGTCCGCCCCGCGAGCACAGAGCCTCGCCTTTGCCGATCCGCCGCCCGTCCACACCCGC CGCCAGCTCACCATGGATGATGATATCGCCGCGCTCGTCGTCGACAACGGCTCCGGCATGTGCAAGGCCGGCTTCGCGGGCGACGATGCC CCCCGGGCCGTCTTCCCCTCCATCGTGGGGCGCCCCAGGCACCAGGGCCTTCCATTCCAGGCCCACCAGGACCCCGAGGCCCACCAGGGG AGGGTTTGCCAGGCCCACCAGGCCCACCAGGATCGTTCCTGTCCAACTCAGAAACCTTCCTCTCCGGCCCCCCAGGCCCACCTGGCCCCC CAGGTCCCAAGGGAGACCAAGGTCCCCCAGGCCCCAGAGGACACCAAGGCGAGCAAGGCCTCCCAGGTTTCTCAACCTCAGGGTCCAGTT CTTTCGGACTCAACCTTCAGGGACCACCAGGCCCACCTGGCCCCCAGGGACCCAAAGGTGACAAAGGTGATCCAGGTGTTCCAGGGGCTC TTGGCATTCCTAGTGGTCCTTCTGAAGGGGGATCATCAAGTACCATGTACGTGTCAGGCCCGCCAGGGCCCCCTGGGCCCCCTGGGCCTC CGGGCTCTATCAGCAGCTCTGGCCAGGAGATTCAGCAGTACATCTCTGAGTACATGCAGAGTGACAGTATTAGATCTTACCTATCCGGAG TTCAGGGTCCCCCAGGCCCACCTGGTCCCCCAGGACCTGTCACCACCATCACAGGCGAGACTTTCGACTACTCAGAGCTGGCAAGCCACG TTGTGAGCTACTTACGGACTTCGGGGTACGGTGTCAGCTTGTTCTCGTCCTCCATCTCTTCTGAAGACATTCTGGCTGTGCTGCAGCGGG ATGACGTGCGTCAGTACCTACGTCAGTACTTGATGGGCCCTCGGGGTCCGCCAGGGCCACCAGGAGCCAGTGGAGATGGGTCCCTCCTGT CTTTGGACTATGCAGAGCTGAGTAGTCGCATTCTCAGCTACATGTCGAGTTCTGGGATCAGCATTGGGCTTCCTGGTCCCCCGGGGCCCC CTGGCTTGCCGGGAACCTCCTATGAGGAGCTCCTCTCCTTGCTGCGAGGGTCTGAATTCAGAGGCATCGTTGGACCCCCAGGTCCCCCGG GTCCACCAGGGATCCCAGGCAATGTGTGGTCCAGCATCAGCGTGGAGGACCTCTCGTCTTACTTACATACTGCCGGCTTGTCATTCATCC CAGGCCCTCCAGGACCTCCTGGTCCCCCAGGGCCTCGAGGGCCCCCGGGTGTCTCAGGAGCCCTGGCAACCTATGCAGCTGAAAACAGCG ACAGCTTCCGGAGCGAGCTGATCAGCTACCTCACAAGTCCTGATGTGCGCAGCTTCATTGTTGGCCCCCCAGGCCCTCCTGGGCCGCAGG GACCCCCTGGGGACAGCCGCCTCCTGTCCACGGATGCCTCCCACAGTCGGGGTAGCAGCTCCTCCTCACACAGCTCATCTGTCAGGCGGG GCAGCTCCTACAGCTCTTCCATGAGCACAGGAGGAGGTGGTGCAGGCTCCCTGGGTGCAGGCGGTGCCTTTGGTGAAGCTGCAGGAGACA GGGGTCCCTATGGCACTGACATCGGCCCAGGCGGAGGCTATGGGGCAGCAGCAGAAGGCGGCATGTATGCTGGCAATGGCGGACTATTGG GAGCTGACTTTGCTGGAGATCTGGATTACAATGAGCTGGCTGTGAGGGTGTCAGAGAGCATGCAGCGTCAGGGCCTACTGCAAGGGATGG CCTACACTGTCCAGGGCCCACCAGGCCAGCCTGGGCCACAGGGGCCACCCGGCATCAGCAAGGTCTTCTCTGCCTACAGCAACGTGACTG CGGACCTCATGGACTTCTTCCAAACTTATGGAGCCATTCAAGGACCCCCTGGGCAAAAAGGAGAGATGGGCACTCCAGGACCCAAAGGTG ACAGGGGCCCTGCTGGGCCACCAGGTCATCCTGGGCCACCTGGCCCTCGAGGACACAAGGGAGAAAAAGGAGACAAAGGTGACCAAGTCT ATGCTGGGCGGAGAAGGAGAAGAAGTATTGCTGTCAAGCCGTGAGCTAGCCATGGCAGGACAGCTCCTGGACCAGGTCTCATAATGCATG TGGCACTTAGGTCCAAGGTCTCCAGAGGGTGAAAGCTGGAGTCTGTCAATGTCCTACTGAGACAGCACAGCCAACCTAGCTAGCAACATT TGTTTTAGTCTGAACAATATATACTTATAGAATTCAGTCAAAGATACACAATCTGAAACAGCTTCATGGGGTGGACTCTAACAGTAGTTG CAATGTTTTAGAATGAGACTTACTTCTCTGCTATCTAGATCTGAACTCCTTGGCTTCTTTACTTAGTTCAAGCCCCAGCCTAGGAAAGCC AGTTACATAAAAGTTGGCTCAGGAGTCTTAGAGCTTTACCTAAATATGAGCCCAGAAAACGGAGGATGGGGGTGGGGCGCCTTCCTGGAG GTGACACTTGATGGGGGTGTGTTCTGGTTACTGTTCTAAGGCTGTGCCATCAGCTCCTTCCTCCCCTGTTCATTCTGCATTCTCTAGTCA GTTGGCTAAGAAGTGACTCTTGCAACTAAAAAAATTAAGAAATTCACTTCCCCTCTAGGAGGTGATGATAGGGTTTCTAATGGTTATATG TATATCACATTCCCATTTGCTTAGAAAGTCTGATTGTAGCTATGATTGTCCGTAGGCCCATACTAGAGTTCATGGATATGTTATACTGAA CCAGGCCAGAGCAAACAGAAAAAGAAGGTTGAGGGCAATGGACAAGGAAGGAATAAAGGGAGAAGAGGGAAAACAGAAAACCTGATGCTG GGGACACAGCATCAGCTCAAGACGTCACCCTCCATTCTGCACTCAGAAAATGGCACTTGGGGGACTGGGCGCAGTTGGTCTTTAACCACT TTTCAATGTCTAAAAACATTTGTTTGTGGTCTATAAGATGAAACATCATTTCAATCGTAAAATTTCCCATTAAAGAAGTTTTTTTTTCTA ATA >1584_1584_1_ACTB-COL17A1_ACTB_chr7_5569166_ENST00000331789_COL17A1_chr10_105801102_ENST00000353479_length(amino acids)=612AA_BP=6 MPGPPGPPGSFLSNSETFLSGPPGPPGPPGPKGDQGPPGPRGHQGEQGLPGFSTSGSSSFGLNLQGPPGPPGPQGPKGDKGDPGVPGALG IPSGPSEGGSSSTMYVSGPPGPPGPPGPPGSISSSGQEIQQYISEYMQSDSIRSYLSGVQGPPGPPGPPGPVTTITGETFDYSELASHVV SYLRTSGYGVSLFSSSISSEDILAVLQRDDVRQYLRQYLMGPRGPPGPPGASGDGSLLSLDYAELSSRILSYMSSSGISIGLPGPPGPPG LPGTSYEELLSLLRGSEFRGIVGPPGPPGPPGIPGNVWSSISVEDLSSYLHTAGLSFIPGPPGPPGPPGPRGPPGVSGALATYAAENSDS FRSELISYLTSPDVRSFIVGPPGPPGPQGPPGDSRLLSTDASHSRGSSSSSHSSSVRRGSSYSSSMSTGGGGAGSLGAGGAFGEAAGDRG PYGTDIGPGGGYGAAAEGGMYAGNGGLLGADFAGDLDYNELAVRVSESMQRQGLLQGMAYTVQGPPGQPGPQGPPGISKVFSAYSNVTAD LMDFFQTYGAIQGPPGQKGEMGTPGPKGDRGPAGPPGHPGPPGPRGHKGEKGDKGDQVYAGRRRRRSIAVKP -------------------------------------------------------------- >1584_1584_2_ACTB-COL17A1_ACTB_chr7_5569166_ENST00000331789_COL17A1_chr10_105801102_ENST00000369733_length(transcript)=2907nt_BP=315nt GAGTGAGCGGCGCGGGGCCAATCAGCGTGCGCCGTTCCGAAAGTTGCCTTTTATGGCTCGAGCGGCCGCGGCGGCGCCCTATAAAACCCA GCGGCGCGACGCGCCACCACCGCCGAGACCGCGTCCGCCCCGCGAGCACAGAGCCTCGCCTTTGCCGATCCGCCGCCCGTCCACACCCGC CGCCAGCTCACCATGGATGATGATATCGCCGCGCTCGTCGTCGACAACGGCTCCGGCATGTGCAAGGCCGGCTTCGCGGGCGACGATGCC CCCCGGGCCGTCTTCCCCTCCATCGTGGGGCGCCCCAGGCACCAGGGCCTTCCATTCCAGGCCCACCAGGACCCCGAGGCCCACCAGGGG AGGGTTTGCCAGGCCCACCAGGCCCACCAGGATCGTTCCTGTCCAACTCAGAAACCTTCCTCTCCGGCCCCCCAGGCCCACCTGGCCCCC CAGGTCCCAAGGGAGACCAAGGTGATCCAGGTGTTCCAGGGGCTCTTGGCATTCCTAGTGGTCCTTCTGAAGGGGGATCATCAAGTACCA TGTACGTGTCAGGCCCGCCAGGGCCCCCTGGGCCCCCTGGGCCTCCGGGCTCTATCAGCAGCTCTGGCCAGGAGATTCAGCAGTACATCT CTGAGTACATGCAGAGTGACAGTATTAGATCTTACCTATCCGGAGTTCAGGGTCCCCCAGGCCCACCTGGTCCCCCAGGACCTGTCACCA CCATCACAGGCGAGACTTTCGACTACTCAGAGCTGGCAAGCCACGTTGTGAGCTACTTACGGACTTCGGGGTACGGTGTCAGCTTGTTCT CGTCCTCCATCTCTTCTGAAGACATTCTGGCTGTGCTGCAGCGGGATGACGTGCGTCAGTACCTACGTCAGTACTTGATGGGCCCTCGGG GTCCGCCAGGGCCACCAGGAGCCAGTGGAGATGGGTCCCTCCTGTCTTTGGACTATGCAGAGCTGAGTAGTCGCATTCTCAGCTACATGT CGAGTTCTGGGATCAGCATTGGGCTTCCTGGTCCCCCGGGGCCCCCTGGCTTGCCGGGAACCTCCTATGAGGAGCTCCTCTCCTTGCTGC GAGCTGCCGGCTTGTCATTCATCCCAGGCCCTCCAGGACCTCCTGGTCCCCCAGGGCCTCGAGGGCCCCCGGGTGTCTCAGGAGCCCTGG CAACCTATGCAGCTGAAAACAGCGACAGCTTCCGGAGCGAGCTGATCAGCTACCTCACAAGTCCTGATGTGCGCAGCTTCATTGTTGGCC CCCCAGGCCCTCCTGGGCCGCAGGGACCCCCTGGGGACAGCCGCCTCCTGTCCACGGATGCCTCCCACAGTCGGGGTAGCAGCTCCTCCT CACACAGCTCATCTGTCAGGCGGGGCAGCTCCTACAGCTCTTCCATGAGCACAGGAGGAGGTGGTGCAGGCTCCCTGGGTGCAGGCGGTG CCTTTGGTGAAGCTGCAGGAGACAGGGGTCCCTATGGCACTGACATCGGCCCAGGCGGAGGCTATGGGGCAGCAGCAGAAGGCGGCATGT ATGCTGGCAATGGCGGACTATTGGGAGCTGACTTTGCTGGAGATCTGGATTACAATGAGCTGGCTGTGAGGGTGTCAGAGAGCATGCAGC GTCAGGGCCTACTGCAAGGGATGGCCTACACTGTCCAGGGCCCACCAGGCCAGCCTGGGCCACAGGGGCCACCCGGCATCAGCAAGGTCT TCTCTGCCTACAGCAACGTGACTGCGGACCTCATGGACTTCTTCCAAACTTATGGAGCCATTCAAGGACCCCCTGGGCAAAAAGGAGAGA TGGGCACTCCAGGACCCAAAGGTGACAGGGGCCCTGCTGGGCCACCAGGTCATCCTGGGCCACCTGGCCCTCGAGGACACAAGGGAGAAA AAGGAGACAAAGGTGACCAAGTCTATGCTGGGCGGAGAAGGAGAAGAAGTATTGCTGTCAAGCCGTGAGCTAGCCATGGCAGGACAGCTC CTGGACCAGGTCTCATAATGCATGTGGCACTTAGGTCCAAGGTCTCCAGAGGGTGAAAGCTGGAGTCTGTCAATGTCCTACTGAGACAGC ACAGCCAACCTAGCTAGCAACATTTGTTTTAGTCTGAACAATATATACTTATAGAATTCAGTCAAAGATACACAATCTGAAACAGCTTCA TGGGGTGGACTCTAACAGTAGTTGCAATGTTTTAGAATGAGACTTACTTCTCTGCTATCTAGATCTGAACTCCTTGGCTTCTTTACTTAG TTCAAGCCCCAGCCTAGGAAAGCCAGTTACATAAAAGTTGGCTCAGGAGTCTTAGAGCTTTACCTAAATATGAGCCCAGAAAACGGAGGA TGGGGGTGGGGCGCCTTCCTGGAGGTGACACTTGATGGGGGTGTGTTCTGGTTACTGTTCTAAGGCTGTGCCATCAGCTCCTTCCTCCCC TGTTCATTCTGCATTCTCTAGTCAGTTGGCTAAGAAGTGACTCTTGCAACTAAAAAAATTAAGAAATTCACTTCCCCTCTAGGAGGTGAT GATAGGGTTTCTAATGGTTATATGTATATCACATTCCCATTTGCTTAGAAAGTCTGATTGTAGCTATGATTGTCCGTAGGCCCATACTAG AGTTCATGGATATGTTATACTGAACCAGGCCAGAGCAAACAGAAAAAGAAGGTTGAGGGCAATGGACAAGGAAGGAATAAAGGGAGAAGA GGGAAAACAGAAAACCTGATGCTGGGGACACAGCATCAGCTCAAGACGTCACCCTCCATTCTGCACTCAGAAAATGGCACTTGGGGGACT GGGCGCAGTTGGTCTTTAACCACTTTTCAATGTCTAAAAACATTTGTTTGTGGTCTATAAGATGAAACATCATTTCAATCGTAAAATTTC CCATTAAAGAAGTTTTTTTTTCTAATA >1584_1584_2_ACTB-COL17A1_ACTB_chr7_5569166_ENST00000331789_COL17A1_chr10_105801102_ENST00000369733_length(amino acids)=530AA_BP=6 MPGPPGPPGSFLSNSETFLSGPPGPPGPPGPKGDQGDPGVPGALGIPSGPSEGGSSSTMYVSGPPGPPGPPGPPGSISSSGQEIQQYISE YMQSDSIRSYLSGVQGPPGPPGPPGPVTTITGETFDYSELASHVVSYLRTSGYGVSLFSSSISSEDILAVLQRDDVRQYLRQYLMGPRGP PGPPGASGDGSLLSLDYAELSSRILSYMSSSGISIGLPGPPGPPGLPGTSYEELLSLLRAAGLSFIPGPPGPPGPPGPRGPPGVSGALAT YAAENSDSFRSELISYLTSPDVRSFIVGPPGPPGPQGPPGDSRLLSTDASHSRGSSSSSHSSSVRRGSSYSSSMSTGGGGAGSLGAGGAF GEAAGDRGPYGTDIGPGGGYGAAAEGGMYAGNGGLLGADFAGDLDYNELAVRVSESMQRQGLLQGMAYTVQGPPGQPGPQGPPGISKVFS AYSNVTADLMDFFQTYGAIQGPPGQKGEMGTPGPKGDRGPAGPPGHPGPPGPRGHKGEKGDKGDQVYAGRRRRRSIAVKP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACTB-COL17A1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACTB-COL17A1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACTB-COL17A1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ACTB | C1855722 | Iris Coloboma with Ptosis, Hypertelorism, and Mental Retardation | 6 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ACTB | C1846331 | Juvenile-onset dystonia | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ACTB | C1853623 | Fryns-Aftimos Syndrome | 2 | GENOMICS_ENGLAND |
| Hgene | ACTB | C2239176 | Liver carcinoma | 2 | CTD_human |
| Hgene | ACTB | C0003129 | Anoxemia | 1 | CTD_human |
| Hgene | ACTB | C0003130 | Anoxia | 1 | CTD_human |
| Hgene | ACTB | C0005586 | Bipolar Disorder | 1 | PSYGENET |
| Hgene | ACTB | C0005818 | Blood Platelet Disorders | 1 | GENOMICS_ENGLAND |
| Hgene | ACTB | C0007097 | Carcinoma | 1 | CTD_human |
| Hgene | ACTB | C0009363 | Congenital ocular coloboma (disorder) | 1 | CTD_human |
| Hgene | ACTB | C0013393 | Dysostoses | 1 | CTD_human |
| Hgene | ACTB | C0013421 | Dystonia | 1 | CTD_human |
| Hgene | ACTB | C0014859 | Esophageal Neoplasms | 1 | CTD_human |
| Hgene | ACTB | C0018784 | Sensorineural Hearing Loss (disorder) | 1 | CTD_human |
| Hgene | ACTB | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Hgene | ACTB | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Hgene | ACTB | C0024667 | Animal Mammary Neoplasms | 1 | CTD_human |
| Hgene | ACTB | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| Hgene | ACTB | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Hgene | ACTB | C0029408 | Degenerative polyarthritis | 1 | CTD_human |
| Hgene | ACTB | C0036341 | Schizophrenia | 1 | PSYGENET |
| Hgene | ACTB | C0086743 | Osteoarthrosis Deformans | 1 | CTD_human |
| Hgene | ACTB | C0151744 | Myocardial Ischemia | 1 | CTD_human |
| Hgene | ACTB | C0205696 | Anaplastic carcinoma | 1 | CTD_human |
| Hgene | ACTB | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human |
| Hgene | ACTB | C0205698 | Undifferentiated carcinoma | 1 | CTD_human |
| Hgene | ACTB | C0205699 | Carcinomatosis | 1 | CTD_human |
| Hgene | ACTB | C0242184 | Hypoxia | 1 | CTD_human |
| Hgene | ACTB | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Hgene | ACTB | C0263579 | Pigmented hairy epidermal nevus | 1 | ORPHANET |
| Hgene | ACTB | C0265541 | Cranioschisis | 1 | CTD_human |
| Hgene | ACTB | C0272183 | Qualitative abnormality of granulocyte | 1 | GENOMICS_ENGLAND |
| Hgene | ACTB | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | ACTB | C0393588 | Dystonia, Paroxysmal | 1 | CTD_human |
| Hgene | ACTB | C0393610 | Dystonia, Diurnal | 1 | CTD_human |
| Hgene | ACTB | C0497552 | Congenital neurologic anomalies | 1 | CTD_human |
| Hgene | ACTB | C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| Hgene | ACTB | C0700292 | Hypoxemia | 1 | CTD_human |
| Hgene | ACTB | C0751093 | Dystonia, Limb | 1 | CTD_human |
| Hgene | ACTB | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Hgene | ACTB | C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human |
| Hgene | ACTB | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Hgene | ACTB | C1691779 | Sensory hearing loss | 1 | CTD_human |
| Hgene | ACTB | C1858042 | Becker Nevus Syndrome | 1 | ORPHANET |
| Hgene | ACTB | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Hgene | ACTB | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Hgene | ACTB | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
| Hgene | ACTB | C4554007 | Uveoretinal Coloboma | 1 | CTD_human |
| Tgene | C0268374 | Adult junctional epidermolysis bullosa (disorder) | 7 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0079301 | Junctional Epidermolysis Bullosa | 2 | GENOMICS_ENGLAND | |
| Tgene | C1852551 | Epithelial Recurrent Erosion Dystrophy | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0002452 | Amelogenesis Imperfecta | 1 | GENOMICS_ENGLAND | |
| Tgene | C2608084 | EPIDERMOLYSIS BULLOSA, JUNCTIONAL, LOCALISATA VARIANT (disorder) | 1 | CTD_human |
