
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ACTG2-ACTA2 (FusionGDB2 ID:HG72TG59) |
Fusion Gene Summary for ACTG2-ACTA2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ACTG2-ACTA2 | Fusion gene ID: hg72tg59 | Hgene | Tgene | Gene symbol | ACTG2 | ACTA2 | Gene ID | 72 | 59 |
| Gene name | actin gamma 2, smooth muscle | actin alpha 2, smooth muscle | |
| Synonyms | ACT|ACTA3|ACTE|ACTL3|ACTSG|VSCM | ACTSA | |
| Cytomap | ('ACTG2')('ACTA2') 2p13.1 | 10q23.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | actin, gamma-enteric smooth muscleactin, gamma 2, smooth muscle, entericactin-like proteinalpha-actin-3 | actin, aortic smooth muscleactin, alpha 2, smooth muscle, aortaalpha-cardiac actincell growth-inhibiting gene 46 protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000345517, ENST00000409624, ENST00000409731, ENST00000409918, | ||
| Fusion gene scores | * DoF score | 9 X 11 X 5=495 | 9 X 11 X 5=495 |
| # samples | 12 | 12 | |
| ** MAII score | log2(12/495*10)=-2.04439411935845 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/495*10)=-2.04439411935845 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ACTG2 [Title/Abstract] AND ACTA2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ACTG2(74128564)-ACTA2(90707143), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ACTG2 (5'-gene) Fusion gene breakpoints across ACTG2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
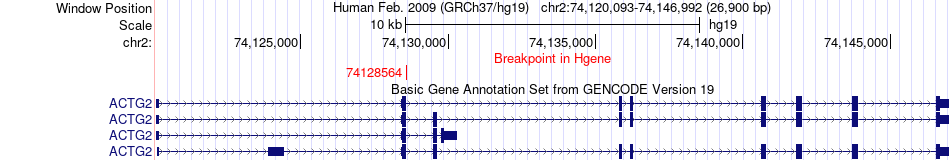 |
 Fusion gene breakpoints across ACTA2 (3'-gene) Fusion gene breakpoints across ACTA2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
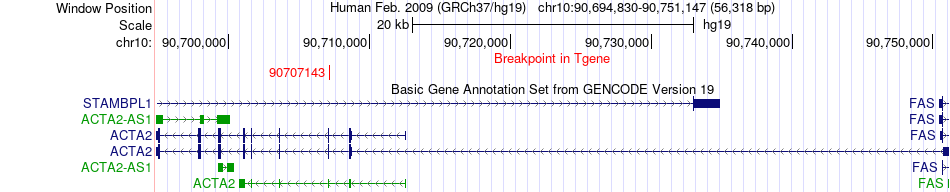 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-FP-7735-11A | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| ChimerDB4 | Non-Cancer | TCGA-HU-A4GC-11A | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
Top |
Fusion Gene ORF analysis for ACTG2-ACTA2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000345517 | ENST00000480297 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| 5CDS-5UTR | ENST00000409624 | ENST00000480297 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| 5CDS-5UTR | ENST00000409731 | ENST00000480297 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| 5CDS-5UTR | ENST00000409918 | ENST00000480297 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000345517 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000345517 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409624 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409624 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409731 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409731 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409918 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
| In-frame | ENST00000409918 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000409731 | ACTG2 | chr2 | 74128564 | + | ENST00000224784 | ACTA2 | chr10 | 90707143 | - | 1401 | 247 | 16 | 1251 | 411 |
| ENST00000409731 | ACTG2 | chr2 | 74128564 | + | ENST00000458208 | ACTA2 | chr10 | 90707143 | - | 1399 | 247 | 16 | 1251 | 411 |
| ENST00000345517 | ACTG2 | chr2 | 74128564 | + | ENST00000224784 | ACTA2 | chr10 | 90707143 | - | 1389 | 235 | 4 | 1239 | 411 |
| ENST00000345517 | ACTG2 | chr2 | 74128564 | + | ENST00000458208 | ACTA2 | chr10 | 90707143 | - | 1387 | 235 | 4 | 1239 | 411 |
| ENST00000409918 | ACTG2 | chr2 | 74128564 | + | ENST00000224784 | ACTA2 | chr10 | 90707143 | - | 1386 | 232 | 1 | 1236 | 411 |
| ENST00000409918 | ACTG2 | chr2 | 74128564 | + | ENST00000458208 | ACTA2 | chr10 | 90707143 | - | 1384 | 232 | 1 | 1236 | 411 |
| ENST00000409624 | ACTG2 | chr2 | 74128564 | + | ENST00000224784 | ACTA2 | chr10 | 90707143 | - | 1923 | 769 | 643 | 1773 | 376 |
| ENST00000409624 | ACTG2 | chr2 | 74128564 | + | ENST00000458208 | ACTA2 | chr10 | 90707143 | - | 1921 | 769 | 643 | 1773 | 376 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000409731 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001359323 | 0.9986407 |
| ENST00000409731 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001402983 | 0.99859697 |
| ENST00000345517 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001335752 | 0.99866426 |
| ENST00000345517 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001373726 | 0.99862623 |
| ENST00000409918 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001383131 | 0.9986168 |
| ENST00000409918 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.001423198 | 0.99857676 |
| ENST00000409624 | ENST00000224784 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.002627882 | 0.99737215 |
| ENST00000409624 | ENST00000458208 | ACTG2 | chr2 | 74128564 | + | ACTA2 | chr10 | 90707143 | - | 0.002710115 | 0.9972899 |
Top |
Fusion Genomic Features for ACTG2-ACTA2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
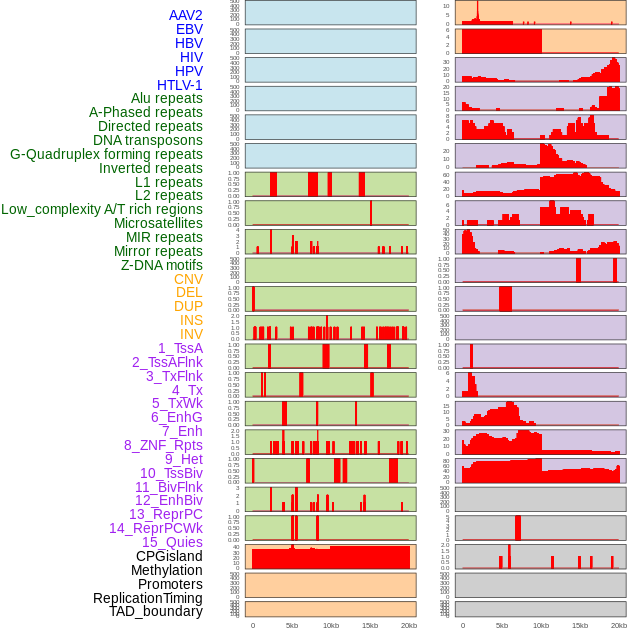 |
Top |
Fusion Protein Features for ACTG2-ACTA2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:74128564/chr10:90707143) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for ACTG2-ACTA2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >1721_1721_1_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000345517_ACTA2_chr10_90707143_ENST00000224784_length(transcript)=1389nt_BP=235nt TATATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCTCCAGTCCCCAGC TCACTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGG AGATGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTA CGTGGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGA AAAGATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAA GGCCAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTA TGCCTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCA TGCCATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTAC TGCTGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTC ATCCTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCA GCCATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCT CTATGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAG CACCATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCA GCAGATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTC TGTCTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACC TATGTGTTTTTTAATAAATCTGAAATAGGCTACTGGTAA >1721_1721_1_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000345517_ACTA2_chr10_90707143_ENST00000224784_length(amino acids)=411AA_BP=37 MLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_2_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000345517_ACTA2_chr10_90707143_ENST00000458208_length(transcript)=1387nt_BP=235nt TATATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCTCCAGTCCCCAGC TCACTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGG AGATGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTA CGTGGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGA AAAGATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAA GGCCAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTA TGCCTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCA TGCCATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTAC TGCTGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTC ATCCTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCA GCCATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCT CTATGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAG CACCATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCA GCAGATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTC TGTCTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACC TATGTGTTTTTTAATAAATCTGAAATAGGCTACTGGT >1721_1721_2_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000345517_ACTA2_chr10_90707143_ENST00000458208_length(amino acids)=411AA_BP=37 MLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_3_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409624_ACTA2_chr10_90707143_ENST00000224784_length(transcript)=1923nt_BP=769nt ACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGGGAATAAAAGCTTCATTCCTGAGTCAGTGAGAGCTTCAATGAAGC CCAGATGTCATTCGTGCTGAAAGAACCAGAACAACTCTCTGCTCCCTGCCAAGCATGAAGCGGTTGTGACCCCAGGAAACCACAGTGACT TTGACTCTGGTTCAGCTGACATGCTCGAGTCTAGCCACAAATTACCAGAAAGCAGCTGAGGAAAGTTTAGAATTTCTCCTGGCAGAGATG GTCCACGAGTCTGGCTCCAGGGTGAACCCCTTCTTCCCAGAGCAGCACGGGGCCCTTTGGAGGCTCGACTCCGCCAGCCCTCCCACTGCC TTCCCCCTGCCTTCCCCACCCTCACTTCGGTTGAGTTTTGCCTTCAAAATAGCAGACTCCAAGCCACCTGAAGGGCTGCAGAACAGCGTT GTTGACAGCTTCAAGTCGCCTTGTGGCTTCTCTGAGCTGCTGCTGTGCTTTGGAAACAAAATTCCAGGCTCTCCCTCTTTCTGTGTGTGG GAAACACCGGAGAGAGTTTGGGGAGGGAGACAAAAGGGCCCATTGAAAACCCAGACAGGCTTGAACTGTGCTCCAGTCCCCAGCTCACTC AGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGGAGATGA TGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTACGTGGG TGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGAAAAGAT CTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAAGGCCAA CCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTATGCCTC TGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCATGCCAT CATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTACTGCTGA GCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTCATCCTC CCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCAGCCATC CTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCTCTATGC TAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAGCACCAT GAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCAGCAGAT GTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTCTGTCTC TAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACCTATGTG TTTTTTAATAAATCTGAAATAGGCTACTGGTAA >1721_1721_3_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409624_ACTA2_chr10_90707143_ENST00000224784_length(amino acids)=376AA_BP=2 MCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHS FYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYASGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLD LAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSSSLEKSYELPDGQVITIGNERFRCPETLFQPSFIGM ESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISK QEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_4_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409624_ACTA2_chr10_90707143_ENST00000458208_length(transcript)=1921nt_BP=769nt ACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGGGAATAAAAGCTTCATTCCTGAGTCAGTGAGAGCTTCAATGAAGC CCAGATGTCATTCGTGCTGAAAGAACCAGAACAACTCTCTGCTCCCTGCCAAGCATGAAGCGGTTGTGACCCCAGGAAACCACAGTGACT TTGACTCTGGTTCAGCTGACATGCTCGAGTCTAGCCACAAATTACCAGAAAGCAGCTGAGGAAAGTTTAGAATTTCTCCTGGCAGAGATG GTCCACGAGTCTGGCTCCAGGGTGAACCCCTTCTTCCCAGAGCAGCACGGGGCCCTTTGGAGGCTCGACTCCGCCAGCCCTCCCACTGCC TTCCCCCTGCCTTCCCCACCCTCACTTCGGTTGAGTTTTGCCTTCAAAATAGCAGACTCCAAGCCACCTGAAGGGCTGCAGAACAGCGTT GTTGACAGCTTCAAGTCGCCTTGTGGCTTCTCTGAGCTGCTGCTGTGCTTTGGAAACAAAATTCCAGGCTCTCCCTCTTTCTGTGTGTGG GAAACACCGGAGAGAGTTTGGGGAGGGAGACAAAAGGGCCCATTGAAAACCCAGACAGGCTTGAACTGTGCTCCAGTCCCCAGCTCACTC AGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGGAGATGA TGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTACGTGGG TGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGAAAAGAT CTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAAGGCCAA CCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTATGCCTC TGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCATGCCAT CATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTACTGCTGA GCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTCATCCTC CCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCAGCCATC CTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCTCTATGC TAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAGCACCAT GAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCAGCAGAT GTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTCTGTCTC TAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACCTATGTG TTTTTTAATAAATCTGAAATAGGCTACTGGT >1721_1721_4_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409624_ACTA2_chr10_90707143_ENST00000458208_length(amino acids)=376AA_BP=2 MCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHS FYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYASGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLD LAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSSSLEKSYELPDGQVITIGNERFRCPETLFQPSFIGM ESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISK QEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_5_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409731_ACTA2_chr10_90707143_ENST00000224784_length(transcript)=1401nt_BP=247nt CCTCTGGGGTTTTATATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCT CCAGTCCCCAGCTCACTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGC AGGCTTCGCAGGAGATGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACA AAAAGACAGCTACGTGGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTG GGACGACATGGAAAAGATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACC CCTGAACCCCAAGGCCAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGT GCTGTCTCTCTATGCCTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTA TGCCTTGCCCCATGCCATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTC CTTCGTTACTACTGCTGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCAC TGCCGCATCCTCATCCTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGA GACCCTGTTCCAGCCATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACAT CAGGAAGGACCTCTATGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGC CCTAGCACCCAGCACCATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCT GTCCACCTTCCAGCAGATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTT CCTGCTCCTCTCTGTCTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTC TGACTCGTTACCTATGTGTTTTTTAATAAATCTGAAATAGGCTACTGGTAA >1721_1721_5_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409731_ACTA2_chr10_90707143_ENST00000224784_length(amino acids)=411AA_BP=37 MLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_6_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409731_ACTA2_chr10_90707143_ENST00000458208_length(transcript)=1399nt_BP=247nt CCTCTGGGGTTTTATATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCT CCAGTCCCCAGCTCACTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGC AGGCTTCGCAGGAGATGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACA AAAAGACAGCTACGTGGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTG GGACGACATGGAAAAGATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACC CCTGAACCCCAAGGCCAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGT GCTGTCTCTCTATGCCTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTA TGCCTTGCCCCATGCCATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTC CTTCGTTACTACTGCTGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCAC TGCCGCATCCTCATCCTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGA GACCCTGTTCCAGCCATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACAT CAGGAAGGACCTCTATGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGC CCTAGCACCCAGCACCATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCT GTCCACCTTCCAGCAGATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTT CCTGCTCCTCTCTGTCTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTC TGACTCGTTACCTATGTGTTTTTTAATAAATCTGAAATAGGCTACTGGT >1721_1721_6_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409731_ACTA2_chr10_90707143_ENST00000458208_length(amino acids)=411AA_BP=37 MLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_7_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409918_ACTA2_chr10_90707143_ENST00000224784_length(transcript)=1386nt_BP=232nt ATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCTCCAGTCCCCAGCTCA CTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGGAGA TGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTACGT GGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGAAAA GATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAAGGC CAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTATGC CTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCATGC CATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTACTGC TGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTCATC CTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCAGCC ATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCTCTA TGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAGCAC CATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCAGCA GATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTCTGT CTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACCTAT GTGTTTTTTAATAAATCTGAAATAGGCTACTGGTAA >1721_1721_7_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409918_ACTA2_chr10_90707143_ENST00000224784_length(amino acids)=411AA_BP=37 LLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- >1721_1721_8_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409918_ACTA2_chr10_90707143_ENST00000458208_length(transcript)=1384nt_BP=232nt ATTGCTCTGGTATTCATGCCAAAGACACACCAGCCCTCAGTCACTGGGAGAAGAACCTCTCATACCCTCGGTGCTCCAGTCCCCAGCTCA CTCAGCCACACACACCATGTGTGAAGAGGAGACCACCGCGCTCGTGTGTGACAATGGCTCTGGCCTGTGCAAGGCAGGCTTCGCAGGAGA TGATGCCCCCCGGGCTGTCTTCCCCTCCATTGTGGGCCGCCCTCGCCACCAGGGGGTGATGGTGGGAATGGGACAAAAAGACAGCTACGT GGGTGACGAAGCACAGAGCAAAAGAGGAATCCTGACCCTGAAGTACCCGATAGAACATGGCATCATCACCAACTGGGACGACATGGAAAA GATCTGGCACCACTCTTTCTACAATGAGCTTCGTGTTGCCCCTGAAGAGCATCCCACCCTGCTCACGGAGGCACCCCTGAACCCCAAGGC CAACCGGGAGAAAATGACTCAAATTATGTTTGAGACTTTCAATGTCCCAGCCATGTATGTGGCTATCCAGGCGGTGCTGTCTCTCTATGC CTCTGGACGCACAACTGGCATCGTGCTGGACTCTGGAGATGGTGTCACCCACAATGTCCCCATCTATGAGGGCTATGCCTTGCCCCATGC CATCATGCGTCTGGATCTGGCTGGCCGAGATCTCACTGACTACCTCATGAAGATCCTGACTGAGCGTGGCTATTCCTTCGTTACTACTGC TGAGCGTGAGATTGTCCGGGACATCAAGGAGAAACTGTGTTATGTAGCTCTGGACTTTGAAAATGAGATGGCCACTGCCGCATCCTCATC CTCCCTTGAGAAGAGTTACGAGTTGCCTGATGGGCAAGTGATCACCATCGGAAATGAACGTTTCCGCTGCCCAGAGACCCTGTTCCAGCC ATCCTTCATCGGGATGGAGTCTGCTGGCATCCATGAAACCACCTACAACAGCATCATGAAGTGTGATATTGACATCAGGAAGGACCTCTA TGCTAACAATGTCCTATCAGGGGGCACCACTATGTACCCTGGCATTGCCGACCGAATGCAGAAGGAGATCACGGCCCTAGCACCCAGCAC CATGAAGATCAAGATCATTGCCCCTCCGGAGCGCAAATACTCTGTCTGGATCGGTGGCTCCATCCTGGCCTCTCTGTCCACCTTCCAGCA GATGTGGATCAGCAAACAGGAATACGATGAAGCCGGGCCTTCCATTGTCCACCGCAAATGCTTCTAAAACACTTTCCTGCTCCTCTCTGT CTCTAGCACACAACTGTGAATGTCCTGTGGAATTATGCCTTCAGTTCTTTTCCAAATCATTCCTAGCCAAAGCTCTGACTCGTTACCTAT GTGTTTTTTAATAAATCTGAAATAGGCTACTGGT >1721_1721_8_ACTG2-ACTA2_ACTG2_chr2_74128564_ENST00000409918_ACTA2_chr10_90707143_ENST00000458208_length(amino acids)=411AA_BP=37 LLWYSCQRHTSPQSLGEEPLIPSVLQSPAHSATHTMCEEETTALVCDNGSGLCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYV GDEAQSKRGILTLKYPIEHGIITNWDDMEKIWHHSFYNELRVAPEEHPTLLTEAPLNPKANREKMTQIMFETFNVPAMYVAIQAVLSLYA SGRTTGIVLDSGDGVTHNVPIYEGYALPHAIMRLDLAGRDLTDYLMKILTERGYSFVTTAEREIVRDIKEKLCYVALDFENEMATAASSS SLEKSYELPDGQVITIGNERFRCPETLFQPSFIGMESAGIHETTYNSIMKCDIDIRKDLYANNVLSGGTTMYPGIADRMQKEITALAPST MKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDEAGPSIVHRKCF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ACTG2-ACTA2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ACTG2-ACTA2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ACTG2-ACTA2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ACTG2 | C0042781 | Visceral Myopathy | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | ACTG2 | C1608393 | Megacystis microcolon intestinal hypoperistalsis syndrome | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | ACTG2 | C0007102 | Malignant tumor of colon | 1 | CTD_human |
| Hgene | ACTG2 | C0009375 | Colonic Neoplasms | 1 | CTD_human |
| Hgene | ACTG2 | C0266833 | Visceral Myopathy, Familial | 1 | CTD_human;ORPHANET |
| Hgene | ACTG2 | C1835084 | Megaduodenum and-or Megacystis | 1 | ORPHANET |
| Tgene | C4707243 | Familial thoracic aortic aneurysm and aortic dissection | 7 | CLINGEN;GENOMICS_ENGLAND | |
| Tgene | C2673186 | Aortic Aneurysm, Familial Thoracic 6 | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C3279690 | MOYAMOYA DISEASE 5 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0023890 | Liver Cirrhosis | 3 | CTD_human | |
| Tgene | C0239946 | Fibrosis, Liver | 3 | CTD_human | |
| Tgene | C3151201 | MULTISYSTEMIC SMOOTH MUSCLE DYSFUNCTION SYNDROME | 3 | GENOMICS_ENGLAND;ORPHANET;UNIPROT | |
| Tgene | C0023893 | Liver Cirrhosis, Experimental | 2 | CTD_human | |
| Tgene | C0026654 | Moyamoya Disease | 2 | GENOMICS_ENGLAND;ORPHANET | |
| Tgene | C2931384 | Moyamoya disease 1 | 2 | ORPHANET | |
| Tgene | C0005398 | Cholestasis, Extrahepatic | 1 | CTD_human | |
| Tgene | C0006142 | Malignant neoplasm of breast | 1 | CTD_human | |
| Tgene | C0007102 | Malignant tumor of colon | 1 | CTD_human | |
| Tgene | C0009375 | Colonic Neoplasms | 1 | CTD_human | |
| Tgene | C0014175 | Endometriosis | 1 | CTD_human | |
| Tgene | C0015934 | Fetal Growth Retardation | 1 | CTD_human | |
| Tgene | C0017668 | Focal glomerulosclerosis | 1 | CTD_human | |
| Tgene | C0019189 | Hepatitis, Chronic | 1 | CTD_human | |
| Tgene | C0022116 | Ischemia | 1 | CTD_human | |
| Tgene | C0022658 | Kidney Diseases | 1 | CTD_human | |
| Tgene | C0023895 | Liver diseases | 1 | CTD_human | |
| Tgene | C0027051 | Myocardial Infarction | 1 | CTD_human | |
| Tgene | C0027719 | Nephrosclerosis | 1 | CTD_human | |
| Tgene | C0034069 | Pulmonary Fibrosis | 1 | CTD_human | |
| Tgene | C0036421 | Systemic Scleroderma | 1 | CTD_human | |
| Tgene | C0041956 | Ureteral obstruction | 1 | CTD_human | |
| Tgene | C0086432 | Hyalinosis, Segmental Glomerular | 1 | CTD_human | |
| Tgene | C0086565 | Liver Dysfunction | 1 | CTD_human | |
| Tgene | C0149519 | Chronic Persistent Hepatitis | 1 | CTD_human | |
| Tgene | C0162872 | Aortic Aneurysm, Thoracic | 1 | CTD_human | |
| Tgene | C0269102 | Endometrioma | 1 | CTD_human | |
| Tgene | C0340630 | Aortic Aneurysm, Thoracoabdominal | 1 | CTD_human | |
| Tgene | C0520463 | Chronic active hepatitis | 1 | CTD_human | |
| Tgene | C0524611 | Cryptogenic Chronic Hepatitis | 1 | CTD_human | |
| Tgene | C0678222 | Breast Carcinoma | 1 | CTD_human | |
| Tgene | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human | |
| Tgene | C1458155 | Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C1619692 | Nephrogenic Fibrosing Dermopathy | 1 | CTD_human | |
| Tgene | C1876165 | Copper-Overload Cirrhosis | 1 | CTD_human | |
| Tgene | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human | |
| Tgene | C4721507 | Alveolitis, Fibrosing | 1 | CTD_human |
