
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CACNA1C-DERA (FusionGDB2 ID:HG775TG51071) |
Fusion Gene Summary for CACNA1C-DERA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CACNA1C-DERA | Fusion gene ID: hg775tg51071 | Hgene | Tgene | Gene symbol | CACNA1C | DERA | Gene ID | 775 | 51071 |
| Gene name | calcium voltage-gated channel subunit alpha1 C | deoxyribose-phosphate aldolase | |
| Synonyms | CACH2|CACN2|CACNL1A1|CCHL1A1|CaV1.2|LQT8|TS|TS. LQT8 | CGI-26|DEOC | |
| Cytomap | ('CACNA1C')('DERA') 12p13.33 | 12p12.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | voltage-dependent L-type calcium channel subunit alpha-1CDHPR, alpha-1 subunitcalcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac musclecalcium channel, cardic dihydropyridine-sensitive, alpha-1 subunitcalcium channel, voltage-dependent, | deoxyribose-phosphate aldolase2-deoxy-D-ribose 5-phosphate aldolase2-deoxyribose-5-phosphate aldolase homologdeoxyriboaldolasedeoxyribose-phosphate aldolase (putative)phosphodeoxyriboaldolaseputative deoxyribose-phosphate aldolase | |
| Modification date | 20200315 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000491104, ENST00000327702, ENST00000335762, ENST00000344100, ENST00000347598, ENST00000399591, ENST00000399595, ENST00000399597, ENST00000399601, ENST00000399603, ENST00000399606, ENST00000399617, ENST00000399621, ENST00000399629, ENST00000399634, ENST00000399637, ENST00000399638, ENST00000399641, ENST00000399644, ENST00000399649, ENST00000399655, ENST00000402845, ENST00000406454, ENST00000480911, | ||
| Fusion gene scores | * DoF score | 19 X 16 X 8=2432 | 10 X 7 X 6=420 |
| # samples | 19 | 9 | |
| ** MAII score | log2(19/2432*10)=-3.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/420*10)=-2.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CACNA1C [Title/Abstract] AND DERA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CACNA1C(2622150)-DERA(16109870), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CACNA1C-DERA seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CACNA1C-DERA seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CACNA1C-DERA seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CACNA1C-DERA seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CACNA1C | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 12130699 |
| Hgene | CACNA1C | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 15454078 |
| Hgene | CACNA1C | GO:0070588 | calcium ion transmembrane transport | 15454078 |
| Tgene | DERA | GO:0046121 | deoxyribonucleoside catabolic process | 9226884 |
 Fusion gene breakpoints across CACNA1C (5'-gene) Fusion gene breakpoints across CACNA1C (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
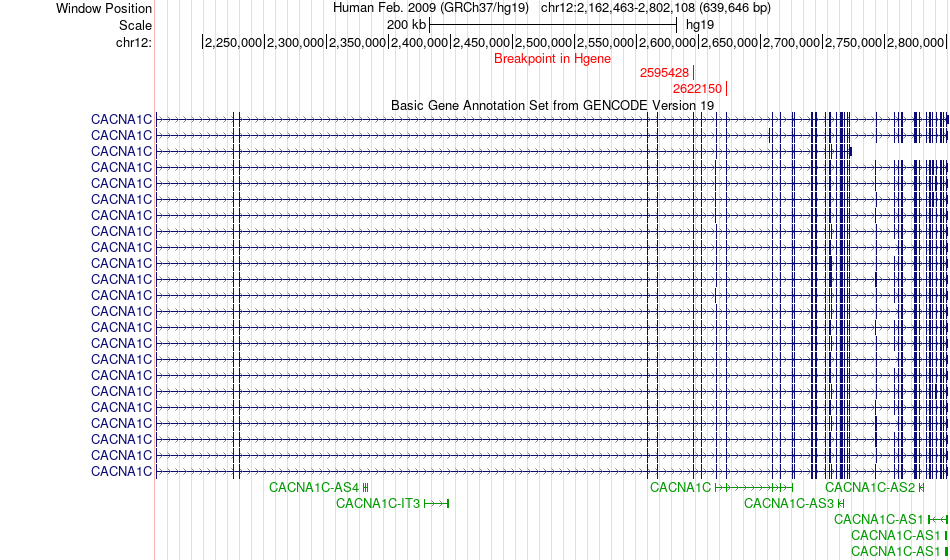 |
 Fusion gene breakpoints across DERA (3'-gene) Fusion gene breakpoints across DERA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
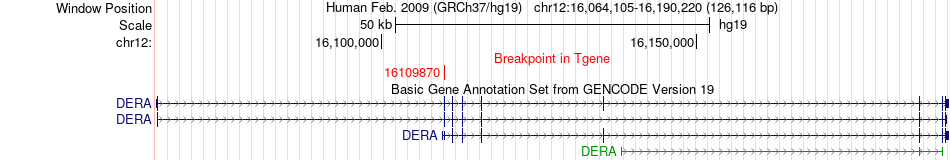 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-EE-A2MR-06A | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| ChimerDB4 | SKCM | TCGA-EE-A2MR-06A | CACNA1C | chr12 | 2622150 | - | DERA | chr12 | 16109870 | + |
| ChimerDB4 | SKCM | TCGA-EE-A2MR-06A | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
Top |
Fusion Gene ORF analysis for CACNA1C-DERA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000491104 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 3UTR-3CDS | ENST00000491104 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 3UTR-5UTR | ENST00000491104 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 3UTR-intron | ENST00000491104 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000327702 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000327702 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000335762 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000335762 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000344100 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000344100 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000347598 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000347598 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399591 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399591 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399595 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399595 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399597 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399597 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399601 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399601 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399603 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399603 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399606 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399606 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399617 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399617 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399621 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399621 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399629 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399629 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399634 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399634 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399637 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399637 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399638 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399638 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399641 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399641 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399644 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399644 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399649 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399649 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399655 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000399655 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000402845 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000402845 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000406454 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000406454 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000480911 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-5UTR | ENST00000480911 | ENST00000526530 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000327702 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000327702 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000335762 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000335762 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000344100 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000344100 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000347598 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000347598 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399591 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399591 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399595 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399595 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399597 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399597 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399601 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399601 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399603 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399603 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399606 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399606 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399617 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399617 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399621 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399621 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399629 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399629 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399634 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399634 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399637 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399637 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399638 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399638 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399641 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399641 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399644 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399644 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399649 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399649 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399655 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000399655 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000402845 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000402845 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000406454 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000406454 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000480911 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| 5CDS-intron | ENST00000480911 | ENST00000532573 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000327702 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000327702 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000327702 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000327702 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000335762 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000335762 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000335762 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000335762 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000344100 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000344100 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000344100 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000344100 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000347598 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000347598 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000347598 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000347598 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399591 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399591 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399591 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399591 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399595 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399595 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399595 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399595 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399597 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399597 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399597 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399597 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399601 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399601 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399601 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399601 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399603 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399603 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399603 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399603 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399606 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399606 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399606 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399606 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399617 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399617 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399617 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399617 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399621 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399621 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399621 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399621 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399629 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399629 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399629 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399629 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399634 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399634 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399634 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399634 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399637 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399637 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399637 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399637 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399638 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399638 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399638 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399638 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399641 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399641 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399641 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399641 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399644 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399644 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399644 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399644 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399649 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399649 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399649 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399649 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399655 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399655 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399655 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000399655 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000402845 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000402845 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000402845 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000402845 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000406454 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000406454 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000406454 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000406454 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000480911 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000480911 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000480911 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| In-frame | ENST00000480911 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + |
| intron-3CDS | ENST00000491104 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| intron-3CDS | ENST00000491104 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| intron-5UTR | ENST00000491104 | ENST00000526530 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
| intron-intron | ENST00000491104 | ENST00000532573 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000335762 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 3121 | 1655 | 265 | 2580 | 771 |
| ENST00000335762 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2457 | 1655 | 265 | 2451 | 728 |
| ENST00000399655 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 3121 | 1655 | 265 | 2580 | 771 |
| ENST00000399655 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2457 | 1655 | 265 | 2451 | 728 |
| ENST00000480911 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2881 | 1415 | 25 | 2340 | 771 |
| ENST00000480911 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2217 | 1415 | 25 | 2211 | 728 |
| ENST00000399644 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399644 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000344100 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000344100 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399638 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399638 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399629 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399629 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399597 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399597 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000327702 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000327702 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399621 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399621 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399649 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399649 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399637 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399637 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000402845 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000402845 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399591 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399591 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399641 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399641 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000347598 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000347598 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399606 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399606 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399595 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399595 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399601 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399601 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399603 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399603 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399634 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399634 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000399617 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000399617 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000406454 | CACNA1C | chr12 | 2622150 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2856 | 1390 | 0 | 2315 | 771 |
| ENST00000406454 | CACNA1C | chr12 | 2622150 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 2192 | 1390 | 0 | 2186 | 728 |
| ENST00000335762 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2647 | 1181 | 265 | 2106 | 613 |
| ENST00000335762 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1983 | 1181 | 265 | 1977 | 570 |
| ENST00000399655 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2647 | 1181 | 265 | 2106 | 613 |
| ENST00000399655 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1983 | 1181 | 265 | 1977 | 570 |
| ENST00000480911 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2407 | 941 | 25 | 1866 | 613 |
| ENST00000480911 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1743 | 941 | 25 | 1737 | 570 |
| ENST00000399644 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399644 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000344100 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000344100 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399638 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399638 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399629 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399629 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399597 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399597 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000327702 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000327702 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399621 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399621 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399649 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399649 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399637 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399637 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000402845 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000402845 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399591 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399591 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399641 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399641 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000347598 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000347598 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399606 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399606 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399595 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399595 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399601 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399601 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399603 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399603 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399634 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399634 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000399617 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000399617 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
| ENST00000406454 | CACNA1C | chr12 | 2595428 | + | ENST00000428559 | DERA | chr12 | 16109870 | + | 2382 | 916 | 0 | 1841 | 613 |
| ENST00000406454 | CACNA1C | chr12 | 2595428 | + | ENST00000532964 | DERA | chr12 | 16109870 | + | 1718 | 916 | 0 | 1712 | 570 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000335762 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003476047 | 0.9965239 |
| ENST00000335762 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.007822551 | 0.9921774 |
| ENST00000399655 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003476047 | 0.9965239 |
| ENST00000399655 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.007822551 | 0.9921774 |
| ENST00000480911 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003792614 | 0.99620736 |
| ENST00000480911 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.008499939 | 0.9915001 |
| ENST00000399644 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399644 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000344100 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000344100 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399638 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399638 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399629 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399629 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399597 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399597 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000327702 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000327702 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399621 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399621 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399649 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399649 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399637 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399637 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000402845 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000402845 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399591 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399591 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399641 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003964947 | 0.99603504 |
| ENST00000399641 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009242802 | 0.99075717 |
| ENST00000347598 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000347598 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399606 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399606 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399595 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399595 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399601 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003951768 | 0.9960483 |
| ENST00000399601 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009111079 | 0.99088895 |
| ENST00000399603 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003964947 | 0.99603504 |
| ENST00000399603 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009242802 | 0.99075717 |
| ENST00000399634 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003964947 | 0.99603504 |
| ENST00000399634 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009242802 | 0.99075717 |
| ENST00000399617 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003964947 | 0.99603504 |
| ENST00000399617 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009242802 | 0.99075717 |
| ENST00000406454 | ENST00000428559 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.003964947 | 0.99603504 |
| ENST00000406454 | ENST00000532964 | CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109870 | + | 0.009242802 | 0.99075717 |
| ENST00000335762 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.004371075 | 0.99562895 |
| ENST00000335762 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.007596398 | 0.9924036 |
| ENST00000399655 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.004371075 | 0.99562895 |
| ENST00000399655 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.007596398 | 0.9924036 |
| ENST00000480911 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.004880865 | 0.9951192 |
| ENST00000480911 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.009186174 | 0.9908138 |
| ENST00000399644 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399644 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000344100 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000344100 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399638 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399638 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399629 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399629 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399597 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399597 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000327702 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000327702 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399621 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399621 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399649 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399649 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399637 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399637 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000402845 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000402845 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399591 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399591 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399641 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399641 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000347598 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000347598 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399606 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399606 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399595 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399595 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399601 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399601 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399603 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399603 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399634 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399634 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000399617 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000399617 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
| ENST00000406454 | ENST00000428559 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.005162625 | 0.99483734 |
| ENST00000406454 | ENST00000532964 | CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109870 | + | 0.010013661 | 0.98998636 |
Top |
Fusion Genomic Features for CACNA1C-DERA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109869 | + | 5.67E-06 | 0.9999943 |
| CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109869 | + | 1.16E-07 | 0.9999999 |
| CACNA1C | chr12 | 2622150 | + | DERA | chr12 | 16109869 | + | 5.67E-06 | 0.9999943 |
| CACNA1C | chr12 | 2595428 | + | DERA | chr12 | 16109869 | + | 1.16E-07 | 0.9999999 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for CACNA1C-DERA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:2622150/chr12:16109870) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 351_372 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 351_372 | 463 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 351_372 | 463 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 351_372 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 351_372 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 351_372 | 463 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 351_372 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 351_372 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 351_372 | 463 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 351_372 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 351_372 | 463 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 351_372 | 463 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 351_372 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 351_372 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 361_364 | 463 | 2174.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 361_364 | 463 | 2180.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 361_364 | 463 | 2187.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 361_364 | 463 | 2147.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 361_364 | 463 | 2147.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 361_364 | 463 | 2159.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 361_364 | 463 | 2174.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 361_364 | 463 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 361_364 | 463 | 2156.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 361_364 | 463 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 361_364 | 463 | 2167.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 361_364 | 463 | 2145.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 361_364 | 463 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 361_364 | 463 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 47_68 | 305 | 2174.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 47_68 | 305 | 2180.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 47_68 | 305 | 2187.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 47_68 | 305 | 2147.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 47_68 | 305 | 2147.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 47_68 | 305 | 2159.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 47_68 | 305 | 2174.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 47_68 | 305 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 47_68 | 305 | 2156.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 47_68 | 305 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 47_68 | 305 | 2167.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 47_68 | 305 | 2145.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 47_68 | 305 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 47_68 | 305 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 47_68 | 463 | 2174.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 47_68 | 463 | 2180.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 47_68 | 463 | 2187.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 47_68 | 463 | 2147.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 47_68 | 463 | 2147.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 47_68 | 463 | 2159.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 47_68 | 463 | 2174.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 47_68 | 463 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 47_68 | 463 | 2156.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 47_68 | 463 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 47_68 | 463 | 2167.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 47_68 | 463 | 2145.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 47_68 | 463 | 2139.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 47_68 | 463 | 2158.0 | Region | Calmodulin-binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 111_408 | 463 | 2174.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 111_408 | 463 | 2180.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 111_408 | 463 | 2187.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 111_408 | 463 | 2147.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 111_408 | 463 | 2147.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 111_408 | 463 | 2159.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 111_408 | 463 | 2174.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 111_408 | 463 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 111_408 | 463 | 2156.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 111_408 | 463 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 111_408 | 463 | 2167.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 111_408 | 463 | 2145.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 111_408 | 463 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 111_408 | 463 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 144_158 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 180_188 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1_124 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 210_232 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 252_268 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 144_158 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 180_188 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1_124 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 210_232 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 252_268 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 144_158 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 180_188 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1_124 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 210_232 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 252_268 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 144_158 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 180_188 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1_124 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 210_232 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 252_268 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 144_158 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 180_188 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1_124 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 210_232 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 252_268 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 144_158 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 180_188 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1_124 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 210_232 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 252_268 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 144_158 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 180_188 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1_124 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 210_232 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 252_268 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 144_158 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 180_188 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1_124 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 210_232 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 252_268 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 144_158 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 180_188 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1_124 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 210_232 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 252_268 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 144_158 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 180_188 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1_124 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 210_232 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 252_268 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 144_158 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 180_188 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1_124 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 210_232 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 252_268 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 144_158 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 180_188 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1_124 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 210_232 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 252_268 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 144_158 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 180_188 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1_124 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 210_232 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 252_268 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 144_158 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 180_188 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1_124 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 210_232 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 252_268 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 144_158 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 180_188 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1_124 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 210_232 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 252_268 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 291_350 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 373_380 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 144_158 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 180_188 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1_124 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 210_232 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 252_268 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 291_350 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 373_380 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 144_158 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 180_188 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1_124 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 210_232 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 252_268 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 291_350 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 373_380 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 144_158 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 180_188 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1_124 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 210_232 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 252_268 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 291_350 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 373_380 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 144_158 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 180_188 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1_124 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 210_232 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 252_268 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 291_350 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 373_380 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 144_158 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 180_188 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1_124 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 210_232 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 252_268 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 291_350 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 373_380 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 144_158 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 180_188 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1_124 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 210_232 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 252_268 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 291_350 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 373_380 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 144_158 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 180_188 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1_124 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 210_232 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 252_268 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 291_350 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 373_380 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 144_158 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 180_188 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1_124 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 210_232 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 252_268 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 291_350 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 373_380 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 144_158 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 180_188 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1_124 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 210_232 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 252_268 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 291_350 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 373_380 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 144_158 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 180_188 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1_124 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 210_232 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 252_268 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 291_350 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 373_380 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 144_158 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 180_188 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1_124 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 210_232 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 252_268 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 291_350 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 373_380 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 144_158 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 180_188 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1_124 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 210_232 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 252_268 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 291_350 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 373_380 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 144_158 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 180_188 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1_124 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 210_232 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 252_268 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 291_350 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 373_380 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 125_143 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 159_179 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 189_209 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 233_251 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 269_290 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 125_143 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 159_179 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 189_209 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 233_251 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 269_290 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 125_143 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 159_179 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 189_209 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 233_251 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 269_290 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 125_143 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 159_179 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 189_209 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 233_251 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 269_290 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 125_143 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 159_179 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 189_209 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 233_251 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 269_290 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 125_143 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 159_179 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 189_209 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 233_251 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 269_290 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 125_143 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 159_179 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 189_209 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 233_251 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 269_290 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 125_143 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 159_179 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 189_209 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 233_251 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 269_290 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 125_143 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 159_179 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 189_209 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 233_251 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 269_290 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 125_143 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 159_179 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 189_209 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 233_251 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 269_290 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 125_143 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 159_179 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 189_209 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 233_251 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 269_290 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 125_143 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 159_179 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 189_209 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 233_251 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 269_290 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 125_143 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 159_179 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 189_209 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 233_251 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 269_290 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 125_143 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 159_179 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 189_209 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 233_251 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 269_290 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 125_143 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 159_179 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 189_209 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 233_251 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 269_290 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 381_401 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 125_143 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 159_179 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 189_209 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 233_251 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 269_290 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 381_401 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 125_143 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 159_179 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 189_209 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 233_251 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 269_290 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 381_401 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 125_143 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 159_179 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 189_209 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 233_251 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 269_290 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 381_401 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 125_143 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 159_179 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 189_209 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 233_251 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 269_290 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 381_401 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 125_143 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 159_179 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 189_209 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 233_251 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 269_290 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 381_401 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 125_143 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 159_179 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 189_209 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 233_251 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 269_290 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 381_401 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 125_143 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 159_179 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 189_209 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 233_251 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 269_290 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 381_401 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 125_143 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 159_179 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 189_209 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 233_251 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 269_290 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 381_401 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 125_143 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 159_179 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 189_209 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 233_251 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 269_290 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 381_401 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 125_143 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 159_179 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 189_209 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 233_251 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 269_290 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 381_401 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 125_143 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 159_179 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 189_209 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 233_251 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 269_290 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 381_401 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 125_143 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 159_179 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 189_209 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 233_251 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 269_290 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 381_401 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 125_143 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 159_179 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 189_209 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 233_251 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 269_290 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat I |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 381_401 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1553_1564 | 305 | 2174.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1553_1564 | 305 | 2180.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1553_1564 | 305 | 2187.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1553_1564 | 305 | 2147.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1553_1564 | 305 | 2147.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1553_1564 | 305 | 2159.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1553_1564 | 305 | 2174.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1553_1564 | 305 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1553_1564 | 305 | 2156.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1553_1564 | 305 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1553_1564 | 305 | 2167.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1553_1564 | 305 | 2145.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1553_1564 | 305 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1553_1564 | 305 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1553_1564 | 463 | 2174.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1553_1564 | 463 | 2180.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1553_1564 | 463 | 2187.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1553_1564 | 463 | 2147.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1553_1564 | 463 | 2147.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1553_1564 | 463 | 2159.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1553_1564 | 463 | 2174.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1553_1564 | 463 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1553_1564 | 463 | 2156.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1553_1564 | 463 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1553_1564 | 463 | 2167.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1553_1564 | 463 | 2145.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1553_1564 | 463 | 2139.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1553_1564 | 463 | 2158.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1167_1173 | 305 | 2174.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 2084_2087 | 305 | 2174.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 654_660 | 305 | 2174.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 768_774 | 305 | 2174.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1167_1173 | 305 | 2180.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 2084_2087 | 305 | 2180.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 654_660 | 305 | 2180.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 768_774 | 305 | 2180.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1167_1173 | 305 | 2187.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 2084_2087 | 305 | 2187.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 654_660 | 305 | 2187.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 768_774 | 305 | 2187.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1167_1173 | 305 | 2147.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 2084_2087 | 305 | 2147.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 654_660 | 305 | 2147.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 768_774 | 305 | 2147.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1167_1173 | 305 | 2147.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 2084_2087 | 305 | 2147.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 654_660 | 305 | 2147.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 768_774 | 305 | 2147.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1167_1173 | 305 | 2159.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 2084_2087 | 305 | 2159.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 654_660 | 305 | 2159.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 768_774 | 305 | 2159.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1167_1173 | 305 | 2174.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 2084_2087 | 305 | 2174.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 654_660 | 305 | 2174.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 768_774 | 305 | 2174.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1167_1173 | 305 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 2084_2087 | 305 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 654_660 | 305 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 768_774 | 305 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1167_1173 | 305 | 2156.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 2084_2087 | 305 | 2156.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 654_660 | 305 | 2156.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 768_774 | 305 | 2156.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1167_1173 | 305 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 2084_2087 | 305 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 654_660 | 305 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 768_774 | 305 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1167_1173 | 305 | 2167.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 2084_2087 | 305 | 2167.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 654_660 | 305 | 2167.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 768_774 | 305 | 2167.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1167_1173 | 305 | 2145.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 2084_2087 | 305 | 2145.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 654_660 | 305 | 2145.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 768_774 | 305 | 2145.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1167_1173 | 305 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 2084_2087 | 305 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 654_660 | 305 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 768_774 | 305 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1167_1173 | 305 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 2084_2087 | 305 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 654_660 | 305 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 768_774 | 305 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1167_1173 | 463 | 2174.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 2084_2087 | 463 | 2174.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 654_660 | 463 | 2174.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 768_774 | 463 | 2174.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1167_1173 | 463 | 2180.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 2084_2087 | 463 | 2180.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 654_660 | 463 | 2180.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 768_774 | 463 | 2180.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1167_1173 | 463 | 2187.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 2084_2087 | 463 | 2187.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 654_660 | 463 | 2187.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 768_774 | 463 | 2187.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1167_1173 | 463 | 2147.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 2084_2087 | 463 | 2147.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 654_660 | 463 | 2147.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 768_774 | 463 | 2147.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1167_1173 | 463 | 2147.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 2084_2087 | 463 | 2147.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 654_660 | 463 | 2147.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 768_774 | 463 | 2147.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1167_1173 | 463 | 2159.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 2084_2087 | 463 | 2159.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 654_660 | 463 | 2159.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 768_774 | 463 | 2159.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1167_1173 | 463 | 2174.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 2084_2087 | 463 | 2174.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 654_660 | 463 | 2174.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 768_774 | 463 | 2174.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1167_1173 | 463 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 2084_2087 | 463 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 654_660 | 463 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 768_774 | 463 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1167_1173 | 463 | 2156.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 2084_2087 | 463 | 2156.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 654_660 | 463 | 2156.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 768_774 | 463 | 2156.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1167_1173 | 463 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 2084_2087 | 463 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 654_660 | 463 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 768_774 | 463 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1167_1173 | 463 | 2167.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 2084_2087 | 463 | 2167.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 654_660 | 463 | 2167.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 768_774 | 463 | 2167.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1167_1173 | 463 | 2145.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 2084_2087 | 463 | 2145.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 654_660 | 463 | 2145.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 768_774 | 463 | 2145.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1167_1173 | 463 | 2139.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 2084_2087 | 463 | 2139.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 654_660 | 463 | 2139.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 768_774 | 463 | 2139.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1167_1173 | 463 | 2158.0 | Compositional bias | Note=Poly-Ile |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 2084_2087 | 463 | 2158.0 | Compositional bias | Note=Poly-Gly |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 654_660 | 463 | 2158.0 | Compositional bias | Note=Poly-Leu |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 768_774 | 463 | 2158.0 | Compositional bias | Note=Poly-Glu |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1122_1142 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1453_1471 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 351_372 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 694_715 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1122_1142 | 305 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1453_1471 | 305 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 351_372 | 305 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 694_715 | 305 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1122_1142 | 305 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1453_1471 | 305 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 351_372 | 305 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 694_715 | 305 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1122_1142 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1453_1471 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 351_372 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 694_715 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1122_1142 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1453_1471 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 351_372 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 694_715 | 305 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1122_1142 | 305 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1453_1471 | 305 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 351_372 | 305 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 694_715 | 305 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1122_1142 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1453_1471 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 351_372 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 694_715 | 305 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1122_1142 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1453_1471 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 351_372 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 694_715 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1122_1142 | 305 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1453_1471 | 305 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 351_372 | 305 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 694_715 | 305 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1122_1142 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1453_1471 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 351_372 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 694_715 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1122_1142 | 305 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1453_1471 | 305 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 351_372 | 305 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 694_715 | 305 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1122_1142 | 305 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1453_1471 | 305 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 351_372 | 305 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 694_715 | 305 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1122_1142 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1453_1471 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 351_372 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 694_715 | 305 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1122_1142 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1453_1471 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 351_372 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 694_715 | 305 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1122_1142 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1453_1471 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 694_715 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1122_1142 | 463 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1453_1471 | 463 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 694_715 | 463 | 2180.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1122_1142 | 463 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1453_1471 | 463 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 694_715 | 463 | 2187.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1122_1142 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1453_1471 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 694_715 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1122_1142 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1453_1471 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 694_715 | 463 | 2147.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1122_1142 | 463 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1453_1471 | 463 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 694_715 | 463 | 2159.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1122_1142 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1453_1471 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 694_715 | 463 | 2174.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1122_1142 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1453_1471 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 694_715 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1122_1142 | 463 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1453_1471 | 463 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 694_715 | 463 | 2156.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1122_1142 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1453_1471 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 694_715 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1122_1142 | 463 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1453_1471 | 463 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 694_715 | 463 | 2167.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1122_1142 | 463 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1453_1471 | 463 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 694_715 | 463 | 2145.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1122_1142 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1453_1471 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 694_715 | 463 | 2139.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1122_1142 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1453_1471 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 694_715 | 463 | 2158.0 | Intramembrane | Pore-forming |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1133_1136 | 305 | 2174.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1462_1465 | 305 | 2174.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 361_364 | 305 | 2174.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 704_707 | 305 | 2174.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1133_1136 | 305 | 2180.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1462_1465 | 305 | 2180.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 361_364 | 305 | 2180.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 704_707 | 305 | 2180.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1133_1136 | 305 | 2187.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1462_1465 | 305 | 2187.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 361_364 | 305 | 2187.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 704_707 | 305 | 2187.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1133_1136 | 305 | 2147.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1462_1465 | 305 | 2147.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 361_364 | 305 | 2147.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 704_707 | 305 | 2147.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1133_1136 | 305 | 2147.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1462_1465 | 305 | 2147.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 361_364 | 305 | 2147.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 704_707 | 305 | 2147.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1133_1136 | 305 | 2159.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1462_1465 | 305 | 2159.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 361_364 | 305 | 2159.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 704_707 | 305 | 2159.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1133_1136 | 305 | 2174.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1462_1465 | 305 | 2174.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 361_364 | 305 | 2174.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 704_707 | 305 | 2174.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1133_1136 | 305 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1462_1465 | 305 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 361_364 | 305 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 704_707 | 305 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1133_1136 | 305 | 2156.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1462_1465 | 305 | 2156.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 361_364 | 305 | 2156.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 704_707 | 305 | 2156.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1133_1136 | 305 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1462_1465 | 305 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 361_364 | 305 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 704_707 | 305 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1133_1136 | 305 | 2167.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1462_1465 | 305 | 2167.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 361_364 | 305 | 2167.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 704_707 | 305 | 2167.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1133_1136 | 305 | 2145.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1462_1465 | 305 | 2145.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 361_364 | 305 | 2145.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 704_707 | 305 | 2145.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1133_1136 | 305 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1462_1465 | 305 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 361_364 | 305 | 2139.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 704_707 | 305 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1133_1136 | 305 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1462_1465 | 305 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 361_364 | 305 | 2158.0 | Motif | Selectivity filter of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 704_707 | 305 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1133_1136 | 463 | 2174.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1462_1465 | 463 | 2174.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 704_707 | 463 | 2174.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1133_1136 | 463 | 2180.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1462_1465 | 463 | 2180.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 704_707 | 463 | 2180.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1133_1136 | 463 | 2187.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1462_1465 | 463 | 2187.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 704_707 | 463 | 2187.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1133_1136 | 463 | 2147.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1462_1465 | 463 | 2147.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 704_707 | 463 | 2147.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1133_1136 | 463 | 2147.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1462_1465 | 463 | 2147.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 704_707 | 463 | 2147.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1133_1136 | 463 | 2159.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1462_1465 | 463 | 2159.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 704_707 | 463 | 2159.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1133_1136 | 463 | 2174.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1462_1465 | 463 | 2174.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 704_707 | 463 | 2174.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1133_1136 | 463 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1462_1465 | 463 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 704_707 | 463 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1133_1136 | 463 | 2156.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1462_1465 | 463 | 2156.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 704_707 | 463 | 2156.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1133_1136 | 463 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1462_1465 | 463 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 704_707 | 463 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1133_1136 | 463 | 2167.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1462_1465 | 463 | 2167.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 704_707 | 463 | 2167.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1133_1136 | 463 | 2145.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1462_1465 | 463 | 2145.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 704_707 | 463 | 2145.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1133_1136 | 463 | 2139.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1462_1465 | 463 | 2139.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 704_707 | 463 | 2139.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1133_1136 | 463 | 2158.0 | Motif | Selectivity filter of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1462_1465 | 463 | 2158.0 | Motif | Selectivity filter of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 704_707 | 463 | 2158.0 | Motif | Selectivity filter of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1109_1198 | 305 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1478_1546 | 305 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1492_1534 | 305 | 2174.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1665_1685 | 305 | 2174.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1699_1718 | 305 | 2174.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1109_1198 | 305 | 2180.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1478_1546 | 305 | 2180.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1492_1534 | 305 | 2180.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1665_1685 | 305 | 2180.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1699_1718 | 305 | 2180.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1109_1198 | 305 | 2187.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1478_1546 | 305 | 2187.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1492_1534 | 305 | 2187.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1665_1685 | 305 | 2187.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1699_1718 | 305 | 2187.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1109_1198 | 305 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1478_1546 | 305 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1492_1534 | 305 | 2147.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1665_1685 | 305 | 2147.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1699_1718 | 305 | 2147.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1109_1198 | 305 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1478_1546 | 305 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1492_1534 | 305 | 2147.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1665_1685 | 305 | 2147.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1699_1718 | 305 | 2147.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1109_1198 | 305 | 2159.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1478_1546 | 305 | 2159.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1492_1534 | 305 | 2159.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1665_1685 | 305 | 2159.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1699_1718 | 305 | 2159.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1109_1198 | 305 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1478_1546 | 305 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1492_1534 | 305 | 2174.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1665_1685 | 305 | 2174.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1699_1718 | 305 | 2174.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1109_1198 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1478_1546 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1492_1534 | 305 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1665_1685 | 305 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1699_1718 | 305 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1109_1198 | 305 | 2156.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1478_1546 | 305 | 2156.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1492_1534 | 305 | 2156.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1665_1685 | 305 | 2156.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1699_1718 | 305 | 2156.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1109_1198 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1478_1546 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1492_1534 | 305 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1665_1685 | 305 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1699_1718 | 305 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1109_1198 | 305 | 2167.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1478_1546 | 305 | 2167.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1492_1534 | 305 | 2167.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1665_1685 | 305 | 2167.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1699_1718 | 305 | 2167.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1109_1198 | 305 | 2145.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1478_1546 | 305 | 2145.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1492_1534 | 305 | 2145.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1665_1685 | 305 | 2145.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1699_1718 | 305 | 2145.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1109_1198 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1478_1546 | 305 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1492_1534 | 305 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1665_1685 | 305 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1699_1718 | 305 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1109_1198 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1478_1546 | 305 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1492_1534 | 305 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1665_1685 | 305 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1699_1718 | 305 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1109_1198 | 463 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1478_1546 | 463 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1492_1534 | 463 | 2174.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1665_1685 | 463 | 2174.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1699_1718 | 463 | 2174.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1109_1198 | 463 | 2180.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1478_1546 | 463 | 2180.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1492_1534 | 463 | 2180.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1665_1685 | 463 | 2180.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1699_1718 | 463 | 2180.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1109_1198 | 463 | 2187.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1478_1546 | 463 | 2187.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1492_1534 | 463 | 2187.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1665_1685 | 463 | 2187.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1699_1718 | 463 | 2187.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1109_1198 | 463 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1478_1546 | 463 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1492_1534 | 463 | 2147.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1665_1685 | 463 | 2147.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1699_1718 | 463 | 2147.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1109_1198 | 463 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1478_1546 | 463 | 2147.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1492_1534 | 463 | 2147.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1665_1685 | 463 | 2147.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1699_1718 | 463 | 2147.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1109_1198 | 463 | 2159.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1478_1546 | 463 | 2159.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1492_1534 | 463 | 2159.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1665_1685 | 463 | 2159.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1699_1718 | 463 | 2159.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1109_1198 | 463 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1478_1546 | 463 | 2174.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1492_1534 | 463 | 2174.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1665_1685 | 463 | 2174.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1699_1718 | 463 | 2174.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1109_1198 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1478_1546 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1492_1534 | 463 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1665_1685 | 463 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1699_1718 | 463 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1109_1198 | 463 | 2156.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1478_1546 | 463 | 2156.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1492_1534 | 463 | 2156.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1665_1685 | 463 | 2156.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1699_1718 | 463 | 2156.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1109_1198 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1478_1546 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1492_1534 | 463 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1665_1685 | 463 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1699_1718 | 463 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1109_1198 | 463 | 2167.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1478_1546 | 463 | 2167.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1492_1534 | 463 | 2167.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1665_1685 | 463 | 2167.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1699_1718 | 463 | 2167.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1109_1198 | 463 | 2145.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1478_1546 | 463 | 2145.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1492_1534 | 463 | 2145.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1665_1685 | 463 | 2145.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1699_1718 | 463 | 2145.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1109_1198 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1478_1546 | 463 | 2139.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1492_1534 | 463 | 2139.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1665_1685 | 463 | 2139.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1699_1718 | 463 | 2139.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1109_1198 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1478_1546 | 463 | 2158.0 | Region | Dihydropyridine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1492_1534 | 463 | 2158.0 | Region | Phenylalkylamine binding |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1665_1685 | 463 | 2158.0 | Region | Calmodulin-binding IQ region |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1699_1718 | 463 | 2158.0 | Region | Important for localization in at the junctional membrane |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 111_408 | 305 | 2174.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1226_1527 | 305 | 2174.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 510_756 | 305 | 2174.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 887_1189 | 305 | 2174.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 111_408 | 305 | 2180.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1226_1527 | 305 | 2180.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 510_756 | 305 | 2180.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 887_1189 | 305 | 2180.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 111_408 | 305 | 2187.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1226_1527 | 305 | 2187.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 510_756 | 305 | 2187.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 887_1189 | 305 | 2187.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 111_408 | 305 | 2147.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1226_1527 | 305 | 2147.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 510_756 | 305 | 2147.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 887_1189 | 305 | 2147.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 111_408 | 305 | 2147.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1226_1527 | 305 | 2147.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 510_756 | 305 | 2147.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 887_1189 | 305 | 2147.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 111_408 | 305 | 2159.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1226_1527 | 305 | 2159.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 510_756 | 305 | 2159.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 887_1189 | 305 | 2159.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 111_408 | 305 | 2174.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1226_1527 | 305 | 2174.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 510_756 | 305 | 2174.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 887_1189 | 305 | 2174.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 111_408 | 305 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1226_1527 | 305 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 510_756 | 305 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 887_1189 | 305 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 111_408 | 305 | 2156.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1226_1527 | 305 | 2156.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 510_756 | 305 | 2156.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 887_1189 | 305 | 2156.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 111_408 | 305 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1226_1527 | 305 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 510_756 | 305 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 887_1189 | 305 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 111_408 | 305 | 2167.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1226_1527 | 305 | 2167.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 510_756 | 305 | 2167.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 887_1189 | 305 | 2167.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 111_408 | 305 | 2145.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1226_1527 | 305 | 2145.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 510_756 | 305 | 2145.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 887_1189 | 305 | 2145.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 111_408 | 305 | 2139.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1226_1527 | 305 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 510_756 | 305 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 887_1189 | 305 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 111_408 | 305 | 2158.0 | Repeat | Note=I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1226_1527 | 305 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 510_756 | 305 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 887_1189 | 305 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1226_1527 | 463 | 2174.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 510_756 | 463 | 2174.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 887_1189 | 463 | 2174.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1226_1527 | 463 | 2180.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 510_756 | 463 | 2180.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 887_1189 | 463 | 2180.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1226_1527 | 463 | 2187.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 510_756 | 463 | 2187.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 887_1189 | 463 | 2187.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1226_1527 | 463 | 2147.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 510_756 | 463 | 2147.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 887_1189 | 463 | 2147.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1226_1527 | 463 | 2147.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 510_756 | 463 | 2147.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 887_1189 | 463 | 2147.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1226_1527 | 463 | 2159.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 510_756 | 463 | 2159.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 887_1189 | 463 | 2159.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1226_1527 | 463 | 2174.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 510_756 | 463 | 2174.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 887_1189 | 463 | 2174.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1226_1527 | 463 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 510_756 | 463 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 887_1189 | 463 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1226_1527 | 463 | 2156.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 510_756 | 463 | 2156.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 887_1189 | 463 | 2156.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1226_1527 | 463 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 510_756 | 463 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 887_1189 | 463 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1226_1527 | 463 | 2167.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 510_756 | 463 | 2167.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 887_1189 | 463 | 2167.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1226_1527 | 463 | 2145.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 510_756 | 463 | 2145.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 887_1189 | 463 | 2145.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1226_1527 | 463 | 2139.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 510_756 | 463 | 2139.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 887_1189 | 463 | 2139.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1226_1527 | 463 | 2158.0 | Repeat | Note=IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 510_756 | 463 | 2158.0 | Repeat | Note=II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 887_1189 | 463 | 2158.0 | Repeat | Note=III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1007_1013 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1033_1051 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1072_1121 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1143_1159 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1182_1239 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1262_1269 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1292_1301 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1322_1372 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1392_1409 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1431_1452 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1472_1499 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1525_2221 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 291_350 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 373_380 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 402_524 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 544_554 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 576_586 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 607_615 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 635_653 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 674_693 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 716_725 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 746_900 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 920_931 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 953_987 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1007_1013 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1033_1051 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1072_1121 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1143_1159 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1182_1239 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1262_1269 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1292_1301 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1322_1372 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1392_1409 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1431_1452 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1472_1499 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1525_2221 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 291_350 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 373_380 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 402_524 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 544_554 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 576_586 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 607_615 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 635_653 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 674_693 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 716_725 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 746_900 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 920_931 | 305 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 953_987 | 305 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1007_1013 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1033_1051 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1072_1121 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1143_1159 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1182_1239 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1262_1269 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1292_1301 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1322_1372 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1392_1409 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1431_1452 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1472_1499 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1525_2221 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 291_350 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 373_380 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 402_524 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 544_554 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 576_586 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 607_615 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 635_653 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 674_693 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 716_725 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 746_900 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 920_931 | 305 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 953_987 | 305 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1007_1013 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1033_1051 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1072_1121 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1143_1159 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1182_1239 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1262_1269 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1292_1301 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1322_1372 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1392_1409 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1431_1452 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1472_1499 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1525_2221 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 291_350 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 373_380 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 402_524 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 544_554 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 576_586 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 607_615 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 635_653 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 674_693 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 716_725 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 746_900 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 920_931 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 953_987 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1007_1013 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1033_1051 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1072_1121 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1143_1159 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1182_1239 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1262_1269 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1292_1301 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1322_1372 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1392_1409 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1431_1452 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1472_1499 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1525_2221 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 291_350 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 373_380 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 402_524 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 544_554 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 576_586 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 607_615 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 635_653 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 674_693 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 716_725 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 746_900 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 920_931 | 305 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 953_987 | 305 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1007_1013 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1033_1051 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1072_1121 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1143_1159 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1182_1239 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1262_1269 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1292_1301 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1322_1372 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1392_1409 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1431_1452 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1472_1499 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1525_2221 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 291_350 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 373_380 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 402_524 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 544_554 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 576_586 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 607_615 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 635_653 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 674_693 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 716_725 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 746_900 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 920_931 | 305 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 953_987 | 305 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1007_1013 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1033_1051 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1072_1121 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1143_1159 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1182_1239 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1262_1269 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1292_1301 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1322_1372 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1392_1409 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1431_1452 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1472_1499 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1525_2221 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 291_350 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 373_380 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 402_524 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 544_554 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 576_586 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 607_615 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 635_653 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 674_693 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 716_725 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 746_900 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 920_931 | 305 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 953_987 | 305 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1007_1013 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1033_1051 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1072_1121 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1143_1159 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1182_1239 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1262_1269 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1292_1301 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1322_1372 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1392_1409 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1431_1452 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1472_1499 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1525_2221 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 291_350 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 373_380 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 402_524 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 544_554 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 576_586 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 607_615 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 635_653 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 674_693 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 716_725 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 746_900 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 920_931 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 953_987 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1007_1013 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1033_1051 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1072_1121 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1143_1159 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1182_1239 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1262_1269 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1292_1301 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1322_1372 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1392_1409 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1431_1452 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1472_1499 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1525_2221 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 291_350 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 373_380 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 402_524 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 544_554 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 576_586 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 607_615 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 635_653 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 674_693 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 716_725 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 746_900 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 920_931 | 305 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 953_987 | 305 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1007_1013 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1033_1051 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1072_1121 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1143_1159 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1182_1239 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1262_1269 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1292_1301 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1322_1372 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1392_1409 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1431_1452 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1472_1499 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1525_2221 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 291_350 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 373_380 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 402_524 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 544_554 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 576_586 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 607_615 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 635_653 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 674_693 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 716_725 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 746_900 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 920_931 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 953_987 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1007_1013 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1033_1051 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1072_1121 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1143_1159 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1182_1239 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1262_1269 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1292_1301 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1322_1372 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1392_1409 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1431_1452 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1472_1499 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1525_2221 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 291_350 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 373_380 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 402_524 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 544_554 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 576_586 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 607_615 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 635_653 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 674_693 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 716_725 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 746_900 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 920_931 | 305 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 953_987 | 305 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1007_1013 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1033_1051 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1072_1121 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1143_1159 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1182_1239 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1262_1269 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1292_1301 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1322_1372 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1392_1409 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1431_1452 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1472_1499 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1525_2221 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 291_350 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 373_380 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 402_524 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 544_554 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 576_586 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 607_615 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 635_653 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 674_693 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 716_725 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 746_900 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 920_931 | 305 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 953_987 | 305 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1007_1013 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1033_1051 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1072_1121 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1143_1159 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1182_1239 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1262_1269 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1292_1301 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1322_1372 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1392_1409 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1431_1452 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1472_1499 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1525_2221 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 291_350 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 373_380 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 402_524 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 544_554 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 576_586 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 607_615 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 635_653 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 674_693 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 716_725 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 746_900 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 920_931 | 305 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 953_987 | 305 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1007_1013 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1033_1051 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1072_1121 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1143_1159 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1182_1239 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1262_1269 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1292_1301 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1322_1372 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1392_1409 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1431_1452 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1472_1499 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1525_2221 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 291_350 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 373_380 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 402_524 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 544_554 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 576_586 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 607_615 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 635_653 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 674_693 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 716_725 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 746_900 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 920_931 | 305 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 953_987 | 305 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1007_1013 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1033_1051 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1072_1121 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1143_1159 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1182_1239 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1262_1269 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1292_1301 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1322_1372 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1392_1409 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1431_1452 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1472_1499 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1525_2221 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 402_524 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 544_554 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 576_586 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 607_615 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 635_653 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 674_693 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 716_725 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 746_900 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 920_931 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 953_987 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1007_1013 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1033_1051 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1072_1121 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1143_1159 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1182_1239 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1262_1269 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1292_1301 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1322_1372 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1392_1409 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1431_1452 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1472_1499 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1525_2221 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 402_524 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 544_554 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 576_586 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 607_615 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 635_653 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 674_693 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 716_725 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 746_900 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 920_931 | 463 | 2180.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 953_987 | 463 | 2180.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1007_1013 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1033_1051 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1072_1121 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1143_1159 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1182_1239 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1262_1269 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1292_1301 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1322_1372 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1392_1409 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1431_1452 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1472_1499 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1525_2221 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 402_524 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 544_554 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 576_586 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 607_615 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 635_653 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 674_693 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 716_725 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 746_900 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 920_931 | 463 | 2187.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 953_987 | 463 | 2187.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1007_1013 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1033_1051 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1072_1121 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1143_1159 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1182_1239 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1262_1269 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1292_1301 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1322_1372 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1392_1409 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1431_1452 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1472_1499 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1525_2221 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 402_524 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 544_554 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 576_586 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 607_615 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 635_653 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 674_693 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 716_725 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 746_900 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 920_931 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 953_987 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1007_1013 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1033_1051 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1072_1121 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1143_1159 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1182_1239 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1262_1269 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1292_1301 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1322_1372 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1392_1409 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1431_1452 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1472_1499 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1525_2221 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 402_524 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 544_554 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 576_586 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 607_615 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 635_653 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 674_693 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 716_725 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 746_900 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 920_931 | 463 | 2147.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 953_987 | 463 | 2147.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1007_1013 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1033_1051 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1072_1121 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1143_1159 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1182_1239 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1262_1269 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1292_1301 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1322_1372 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1392_1409 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1431_1452 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1472_1499 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1525_2221 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 402_524 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 544_554 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 576_586 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 607_615 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 635_653 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 674_693 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 716_725 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 746_900 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 920_931 | 463 | 2159.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 953_987 | 463 | 2159.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1007_1013 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1033_1051 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1072_1121 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1143_1159 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1182_1239 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1262_1269 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1292_1301 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1322_1372 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1392_1409 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1431_1452 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1472_1499 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1525_2221 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 402_524 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 544_554 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 576_586 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 607_615 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 635_653 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 674_693 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 716_725 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 746_900 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 920_931 | 463 | 2174.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 953_987 | 463 | 2174.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1007_1013 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1033_1051 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1072_1121 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1143_1159 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1182_1239 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1262_1269 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1292_1301 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1322_1372 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1392_1409 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1431_1452 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1472_1499 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1525_2221 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 402_524 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 544_554 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 576_586 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 607_615 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 635_653 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 674_693 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 716_725 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 746_900 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 920_931 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 953_987 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1007_1013 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1033_1051 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1072_1121 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1143_1159 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1182_1239 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1262_1269 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1292_1301 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1322_1372 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1392_1409 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1431_1452 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1472_1499 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1525_2221 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 402_524 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 544_554 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 576_586 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 607_615 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 635_653 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 674_693 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 716_725 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 746_900 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 920_931 | 463 | 2156.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 953_987 | 463 | 2156.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1007_1013 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1033_1051 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1072_1121 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1143_1159 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1182_1239 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1262_1269 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1292_1301 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1322_1372 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1392_1409 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1431_1452 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1472_1499 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1525_2221 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 402_524 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 544_554 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 576_586 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 607_615 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 635_653 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 674_693 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 716_725 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 746_900 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 920_931 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 953_987 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1007_1013 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1033_1051 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1072_1121 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1143_1159 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1182_1239 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1262_1269 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1292_1301 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1322_1372 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1392_1409 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1431_1452 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1472_1499 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1525_2221 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 402_524 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 544_554 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 576_586 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 607_615 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 635_653 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 674_693 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 716_725 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 746_900 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 920_931 | 463 | 2167.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 953_987 | 463 | 2167.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1007_1013 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1033_1051 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1072_1121 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1143_1159 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1182_1239 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1262_1269 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1292_1301 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1322_1372 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1392_1409 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1431_1452 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1472_1499 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1525_2221 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 402_524 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 544_554 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 576_586 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 607_615 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 635_653 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 674_693 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 716_725 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 746_900 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 920_931 | 463 | 2145.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 953_987 | 463 | 2145.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1007_1013 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1033_1051 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1072_1121 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1143_1159 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1182_1239 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1262_1269 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1292_1301 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1322_1372 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1392_1409 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1431_1452 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1472_1499 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1525_2221 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 402_524 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 544_554 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 576_586 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 607_615 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 635_653 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 674_693 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 716_725 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 746_900 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 920_931 | 463 | 2139.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 953_987 | 463 | 2139.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1007_1013 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1033_1051 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1072_1121 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1143_1159 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1182_1239 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1262_1269 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1292_1301 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1322_1372 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1392_1409 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1431_1452 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1472_1499 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1525_2221 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 402_524 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 544_554 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 576_586 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 607_615 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 635_653 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 674_693 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 716_725 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 746_900 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 920_931 | 463 | 2158.0 | Topological domain | Extracellular |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 953_987 | 463 | 2158.0 | Topological domain | Cytoplasmic |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1014_1032 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1052_1071 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1160_1181 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1240_1261 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1270_1291 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1302_1321 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1373_1391 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1410_1430 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 1500_1524 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 381_401 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 525_543 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 555_575 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 587_606 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 616_634 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 654_673 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 726_745 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 901_919 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 932_952 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 988_1006 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1014_1032 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1052_1071 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1160_1181 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1240_1261 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1270_1291 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1302_1321 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1373_1391 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1410_1430 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 1500_1524 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 381_401 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 525_543 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 555_575 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 587_606 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 616_634 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 654_673 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 726_745 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 901_919 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 932_952 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 988_1006 | 305 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1014_1032 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1052_1071 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1160_1181 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1240_1261 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1270_1291 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1302_1321 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1373_1391 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1410_1430 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 1500_1524 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 381_401 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 525_543 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 555_575 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 587_606 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 616_634 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 654_673 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 726_745 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 901_919 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 932_952 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 988_1006 | 305 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1014_1032 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1052_1071 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1160_1181 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1240_1261 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1270_1291 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1302_1321 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1373_1391 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1410_1430 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 1500_1524 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 381_401 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 525_543 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 555_575 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 587_606 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 616_634 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 654_673 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 726_745 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 901_919 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 932_952 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 988_1006 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1014_1032 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1052_1071 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1160_1181 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1240_1261 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1270_1291 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1302_1321 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1373_1391 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1410_1430 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 1500_1524 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 381_401 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 525_543 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 555_575 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 587_606 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 616_634 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 654_673 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 726_745 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 901_919 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 932_952 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 988_1006 | 305 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1014_1032 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1052_1071 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1160_1181 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1240_1261 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1270_1291 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1302_1321 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1373_1391 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1410_1430 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 1500_1524 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 381_401 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 525_543 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 555_575 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 587_606 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 616_634 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 654_673 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 726_745 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 901_919 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 932_952 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 988_1006 | 305 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1014_1032 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1052_1071 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1160_1181 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1240_1261 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1270_1291 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1302_1321 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1373_1391 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1410_1430 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 1500_1524 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 381_401 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 525_543 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 555_575 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 587_606 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 616_634 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 654_673 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 726_745 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 901_919 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 932_952 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 988_1006 | 305 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1014_1032 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1052_1071 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1160_1181 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1240_1261 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1270_1291 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1302_1321 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1373_1391 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1410_1430 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 1500_1524 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 381_401 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 525_543 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 555_575 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 587_606 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 616_634 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 654_673 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 726_745 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 901_919 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 932_952 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 988_1006 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1014_1032 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1052_1071 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1160_1181 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1240_1261 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1270_1291 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1302_1321 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1373_1391 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1410_1430 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 1500_1524 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 381_401 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 525_543 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 555_575 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 587_606 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 616_634 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 654_673 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 726_745 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 901_919 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 932_952 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 988_1006 | 305 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1014_1032 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1052_1071 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1160_1181 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1240_1261 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1270_1291 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1302_1321 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1373_1391 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1410_1430 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 1500_1524 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 381_401 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 525_543 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 555_575 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 587_606 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 616_634 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 654_673 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 726_745 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 901_919 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 932_952 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 988_1006 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1014_1032 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1052_1071 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1160_1181 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1240_1261 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1270_1291 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1302_1321 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1373_1391 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1410_1430 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 1500_1524 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 381_401 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 525_543 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 555_575 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 587_606 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 616_634 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 654_673 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 726_745 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 901_919 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 932_952 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 988_1006 | 305 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1014_1032 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1052_1071 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1160_1181 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1240_1261 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1270_1291 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1302_1321 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1373_1391 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1410_1430 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 1500_1524 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 381_401 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 525_543 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 555_575 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 587_606 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 616_634 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 654_673 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 726_745 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 901_919 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 932_952 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 988_1006 | 305 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1014_1032 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1052_1071 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1160_1181 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1240_1261 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1270_1291 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1302_1321 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1373_1391 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1410_1430 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 1500_1524 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 381_401 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 525_543 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 555_575 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 587_606 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 616_634 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 654_673 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 726_745 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 901_919 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 932_952 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 988_1006 | 305 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1014_1032 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1052_1071 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1160_1181 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1240_1261 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1270_1291 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1302_1321 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1373_1391 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1410_1430 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 1500_1524 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 381_401 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat I |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 525_543 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 555_575 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 587_606 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 616_634 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 654_673 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 726_745 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 901_919 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 932_952 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 988_1006 | 305 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1014_1032 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1052_1071 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1160_1181 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1240_1261 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1270_1291 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1302_1321 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1373_1391 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1410_1430 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 1500_1524 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 525_543 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 555_575 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 587_606 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 616_634 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 654_673 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 726_745 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 901_919 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 932_952 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 988_1006 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1014_1032 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1052_1071 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1160_1181 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1240_1261 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1270_1291 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1302_1321 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1373_1391 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1410_1430 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 1500_1524 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 525_543 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 555_575 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 587_606 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 616_634 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 654_673 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 726_745 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 901_919 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 932_952 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 988_1006 | 463 | 2180.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1014_1032 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1052_1071 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1160_1181 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1240_1261 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1270_1291 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1302_1321 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1373_1391 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1410_1430 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 1500_1524 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 525_543 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 555_575 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 587_606 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 616_634 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 654_673 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 726_745 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 901_919 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 932_952 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 988_1006 | 463 | 2187.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1014_1032 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1052_1071 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1160_1181 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1240_1261 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1270_1291 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1302_1321 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1373_1391 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1410_1430 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 1500_1524 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 525_543 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 555_575 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 587_606 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 616_634 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 654_673 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 726_745 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 901_919 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 932_952 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 988_1006 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1014_1032 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1052_1071 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1160_1181 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1240_1261 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1270_1291 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1302_1321 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1373_1391 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1410_1430 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 1500_1524 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 525_543 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 555_575 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 587_606 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 616_634 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 654_673 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 726_745 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 901_919 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 932_952 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 988_1006 | 463 | 2147.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1014_1032 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1052_1071 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1160_1181 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1240_1261 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1270_1291 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1302_1321 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1373_1391 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1410_1430 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 1500_1524 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 525_543 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 555_575 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 587_606 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 616_634 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 654_673 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 726_745 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 901_919 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 932_952 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 988_1006 | 463 | 2159.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1014_1032 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1052_1071 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1160_1181 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1240_1261 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1270_1291 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1302_1321 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1373_1391 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1410_1430 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 1500_1524 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 525_543 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 555_575 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 587_606 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 616_634 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 654_673 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 726_745 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 901_919 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 932_952 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 988_1006 | 463 | 2174.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1014_1032 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1052_1071 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1160_1181 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1240_1261 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1270_1291 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1302_1321 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1373_1391 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1410_1430 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 1500_1524 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 525_543 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 555_575 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 587_606 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 616_634 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 654_673 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 726_745 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 901_919 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 932_952 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 988_1006 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1014_1032 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1052_1071 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1160_1181 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1240_1261 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1270_1291 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1302_1321 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1373_1391 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1410_1430 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 1500_1524 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 525_543 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 555_575 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 587_606 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 616_634 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 654_673 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 726_745 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 901_919 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 932_952 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 988_1006 | 463 | 2156.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1014_1032 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1052_1071 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1160_1181 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1240_1261 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1270_1291 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1302_1321 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1373_1391 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1410_1430 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 1500_1524 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 525_543 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 555_575 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 587_606 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 616_634 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 654_673 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 726_745 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 901_919 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 932_952 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 988_1006 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1014_1032 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1052_1071 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1160_1181 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1240_1261 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1270_1291 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1302_1321 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1373_1391 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1410_1430 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 1500_1524 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 525_543 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 555_575 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 587_606 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 616_634 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 654_673 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 726_745 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 901_919 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 932_952 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 988_1006 | 463 | 2167.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1014_1032 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1052_1071 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1160_1181 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1240_1261 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1270_1291 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1302_1321 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1373_1391 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1410_1430 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 1500_1524 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 525_543 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 555_575 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 587_606 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 616_634 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 654_673 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 726_745 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 901_919 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 932_952 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 988_1006 | 463 | 2145.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1014_1032 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1052_1071 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1160_1181 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1240_1261 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1270_1291 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1302_1321 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1373_1391 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1410_1430 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 1500_1524 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 525_543 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 555_575 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 587_606 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 616_634 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 654_673 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 726_745 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 901_919 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 932_952 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 988_1006 | 463 | 2139.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1014_1032 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1052_1071 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1160_1181 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1240_1261 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1270_1291 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1302_1321 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1373_1391 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1410_1430 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 1500_1524 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat IV |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 525_543 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 555_575 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 587_606 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 616_634 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS4 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 654_673 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS5 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 726_745 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS6 of repeat II |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 901_919 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS1 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 932_952 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS2 of repeat III |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 988_1006 | 463 | 2158.0 | Transmembrane | Helical%3B Name%3DS3 of repeat III |
Top |
Fusion Gene Sequence for CACNA1C-DERA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >12299_12299_1_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000327702_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_1_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000327702_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_2_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000327702_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_2_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000327702_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_3_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000335762_DERA_chr12_16109870_ENST00000428559_length(transcript)=2647nt_BP=1181nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAAC CGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAA CATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGC CGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGC TGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGT AATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCTCATCT TAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGATTTTAT TAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAAC TGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGG AGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGG AAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTT TCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGA ATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTT GTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAAC CGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATT TCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTCATATA CAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_3_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000335762_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_4_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000335762_DERA_chr12_16109870_ENST00000532964_length(transcript)=1983nt_BP=1181nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAAC CGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAA CATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGC CGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGC TGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGT AATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTT CCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCG CAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGC CAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAAT CAG >12299_12299_4_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000335762_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_5_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000344100_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_5_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000344100_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_6_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000344100_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_6_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000344100_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_7_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000347598_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_7_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000347598_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_8_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000347598_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_8_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000347598_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_9_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399591_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_9_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399591_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_10_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399591_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_10_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399591_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_11_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399595_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_11_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399595_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_12_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399595_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_12_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399595_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_13_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399597_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_13_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399597_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_14_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399597_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_14_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399597_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_15_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399601_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_15_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399601_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_16_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399601_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_16_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399601_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_17_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399603_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_17_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399603_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_18_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399603_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_18_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399603_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_19_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399606_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_19_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399606_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_20_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399606_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_20_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399606_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_21_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399617_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_21_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399617_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_22_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399617_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_22_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399617_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_23_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399621_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_23_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399621_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_24_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399621_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_24_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399621_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_25_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399629_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_25_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399629_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_26_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399629_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_26_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399629_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_27_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399634_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_27_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399634_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_28_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399634_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_28_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399634_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_29_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399637_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_29_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399637_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_30_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399637_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_30_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399637_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_31_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399638_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_31_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399638_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_32_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399638_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_32_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399638_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_33_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399641_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_33_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399641_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_34_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399641_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_34_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399641_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_35_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399644_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_35_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399644_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_36_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399644_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_36_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399644_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_37_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399649_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_37_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399649_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_38_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399649_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_38_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399649_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_39_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399655_DERA_chr12_16109870_ENST00000428559_length(transcript)=2647nt_BP=1181nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAAC CGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAA CATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGC CGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGC TGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGT AATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCTCATCT TAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGATTTTAT TAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAAC TGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGG AGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGG AAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTT TCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGA ATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTT GTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAAC CGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATT TCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTCATATA CAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_39_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399655_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_40_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399655_DERA_chr12_16109870_ENST00000532964_length(transcript)=1983nt_BP=1181nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAAC CGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAA CATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGC CGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGC TGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGT AATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTT CCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCG CAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGC CAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAAT CAG >12299_12299_40_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000399655_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_41_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000402845_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_41_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000402845_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_42_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000402845_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_42_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000402845_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_43_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000406454_DERA_chr12_16109870_ENST00000428559_length(transcript)=2382nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCT CATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGAT TTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGG AAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAG CTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTG ACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATT ATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTA ATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTA ACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATAT TAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCAC TAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTC ATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_43_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000406454_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_44_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000406454_DERA_chr12_16109870_ENST00000532964_length(transcript)=1718nt_BP=916nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGC AGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCT TCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACT ACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTG GCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGAC GTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCC ACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGC ATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATA GGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCT TAAATCAG >12299_12299_44_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000406454_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_45_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000480911_DERA_chr12_16109870_ENST00000428559_length(transcript)=2407nt_BP=941nt CTCCTATTAAAACCATTTTTGGTCCATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAG CCCACGCCCCGCCCATGCCAACATGAATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTG GCAGGCGGCCATCGACGCAGCCCGGCAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAA GCGGCAGCAATATGGGAAACCCAAGAAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCC CATCCGGAGGGCCTGCATCAGCATTGTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGC GATCTATATTCCCTTTCCAGAAGATGATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGT GGAAGCGTTTTTAAAAGTAATCGCCTATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAAT TGTGGTTGTGGGGCTTTTTAGTGCAATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGA TGTGAAGGCGCTGAGGGCCTTCCGCGTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCAT CAAGGCCATGGTCCCCCTGCTGCACATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGG GAAGATGCACAAGACCTGCTACAACCAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAG GCGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTAC TACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTT AAATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGG CTGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGT GGAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCA GTTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTAT GATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCT GCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGC TTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACAT TGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAG TTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCT CTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTT AATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAA ATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACG ACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTG TCCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_45_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000480911_DERA_chr12_16109870_ENST00000428559_length(amino acids)=613AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEALYDEIRQFRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFW KTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_46_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000480911_DERA_chr12_16109870_ENST00000532964_length(transcript)=1743nt_BP=941nt CTCCTATTAAAACCATTTTTGGTCCATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAG CCCACGCCCCGCCCATGCCAACATGAATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTG GCAGGCGGCCATCGACGCAGCCCGGCAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAA GCGGCAGCAATATGGGAAACCCAAGAAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCC CATCCGGAGGGCCTGCATCAGCATTGTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGC GATCTATATTCCCTTTCCAGAAGATGATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGT GGAAGCGTTTTTAAAAGTAATCGCCTATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAAT TGTGGTTGTGGGGCTTTTTAGTGCAATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGA TGTGAAGGCGCTGAGGGCCTTCCGCGTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCAT CAAGGCCATGGTCCCCCTGCTGCACATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGG GAAGATGCACAAGACCTGCTACAACCAGGAGGGCATAGCAGACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAG GCGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTAC TACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTT AAATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGG CTGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGT GGAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTC TACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAA GATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTG GCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGC AGCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_46_CACNA1C-DERA_CACNA1C_chr12_2595428_ENST00000480911_DERA_chr12_16109870_ENST00000532964_length(amino acids)=570AA_BP=305 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTS SNIQRLCYKAKYPIREDLLKALNMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEID VVINRSLVLTGQWEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRI GASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_47_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000327702_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_47_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000327702_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_48_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000327702_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_48_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000327702_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_49_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000335762_DERA_chr12_16109870_ENST00000428559_length(transcript)=3121nt_BP=1655nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACGGTGTG CAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATCACCAT GGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATCATAGG GTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGATTTCCA GAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAGAATGA GGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGC GGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACT TTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATAT GCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAA TATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGA TGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCG CAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGC AATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGC CATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCT CTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAG GCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTT ACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTT AAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTAC TAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTA AGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCA GGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAA TGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_49_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000335762_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_50_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000335762_DERA_chr12_16109870_ENST00000532964_length(transcript)=2457nt_BP=1655nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACGGTGTG CAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATCACCAT GGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATCATAGG GTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGATTTCCA GAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAGAATGA GGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGC GGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACT TTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATAT GCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAA TATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGA TGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGG AAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGG GTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAA GCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTA TCATGATCTTCCAATGTCTTAAATCAG >12299_12299_50_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000335762_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_51_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000344100_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_51_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000344100_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_52_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000344100_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_52_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000344100_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_53_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000347598_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_53_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000347598_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_54_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000347598_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_54_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000347598_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_55_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399591_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_55_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399591_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_56_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399591_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_56_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399591_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_57_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399595_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_57_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399595_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_58_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399595_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_58_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399595_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_59_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399597_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_59_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399597_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_60_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399597_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_60_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399597_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_61_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399601_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_61_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399601_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_62_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399601_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_62_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399601_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_63_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399603_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_63_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399603_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_64_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399603_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_64_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399603_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_65_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399606_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_65_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399606_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_66_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399606_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_66_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399606_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_67_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399617_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_67_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399617_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_68_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399617_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_68_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399617_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_69_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399621_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_69_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399621_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_70_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399621_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_70_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399621_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_71_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399629_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_71_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399629_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_72_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399629_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_72_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399629_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_73_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399634_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_73_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399634_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_74_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399634_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_74_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399634_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_75_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399637_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_75_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399637_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_76_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399637_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_76_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399637_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_77_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399638_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_77_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399638_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_78_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399638_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_78_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399638_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_79_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399641_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_79_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399641_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_80_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399641_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_80_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399641_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_81_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399644_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_81_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399644_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_82_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399644_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_82_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399644_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_83_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399649_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_83_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399649_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_84_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399649_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_84_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399649_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_85_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399655_DERA_chr12_16109870_ENST00000428559_length(transcript)=3121nt_BP=1655nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACGGTGTG CAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATCACCAT GGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATCATAGG GTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGATTTCCA GAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAGAATGA GGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGC GGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACT TTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATAT GCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAA TATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGA TGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAGTTTCG CAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATGATAGC AATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGC CATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCT CTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAG GCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTT ACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTT AAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTAC TAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTA AGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCA GGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAA TGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_85_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399655_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_86_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399655_DERA_chr12_16109870_ENST00000532964_length(transcript)=2457nt_BP=1655nt GGAAGGGGCCCGGATGTACTGAGGATGCGTTACAGTTTCACTCGAGGAGGCAGTAGTGGAAAGGAGCAGTTTTTGGGGTTTGATGCCATA ATGGGAATCAGGTAATCGTCGGCGGGGAAGAAGAAACGCTGCAGACCACGGCTTCCTCGAATCTTGCGCGAAAGCCGCCGGCCTCGGAGG AGGGATTAATCCAGACCCGCCGGGGGGTGTTTTCACATTTCTTCCTCTTCGTGGCTGCTCCTCCTATTAAAACCATTTTTGGTCCATGGT CAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATGAATGC CAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGGCAGGC TAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAGAAGCA GGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATTGTCGA ATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGATGATTC CAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCCTATGG ACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCAATTTT AGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGCGTGCT GCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCACATCGC CCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAACCAGGA GGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACGGTGTG CAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATCACCAT GGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATCATAGG GTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGATTTCCA GAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAGAATGA GGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGGCGTGC GGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACTACACT TTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTAAATAT GCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGCTGTAA TATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTGGAAGA TGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCTACTGG AAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGG GTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAA GCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTA TCATGATCTTCCAATGTCTTAAATCAG >12299_12299_86_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000399655_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_87_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000402845_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_87_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000402845_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_88_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000402845_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTGGATCTATTTTGTTACACTAATCATC ATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_88_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000402845_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_89_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000406454_DERA_chr12_16109870_ENST00000428559_length(transcript)=2856nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGCCCTGTATGATGAGATTCGTCAG TTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCTTACTAATGTCTATAAAGCCAGTATG ATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTG CGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCT TGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATT GAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGT TCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTC TCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTA ATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAA TTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGA CTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGT CCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAAA >12299_12299_89_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000406454_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_90_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000406454_DERA_chr12_16109870_ENST00000532964_length(transcript)=2192nt_BP=1390nt ATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAGCCCACGCCCCGCCCATGCCAACATG AATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTGGCAGGCGGCCATCGACGCAGCCCGG CAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAAGCGGCAGCAATATGGGAAACCCAAG AAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCCCATCCGGAGGGCCTGCATCAGCATT GTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGCGATCTATATTCCCTTTCCAGAAGAT GATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGTGGAAGCGTTTTTAAAAGTAATCGCC TATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAATTGTGGTTGTGGGGCTTTTTAGTGCA ATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGATGTGAAGGCGCTGAGGGCCTTCCGC GTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCATCAAGGCCATGGTCCCCCTGCTGCAC ATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGGGAAGATGCACAAGACCTGCTACAAC CAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCACGGGCGGCAGTGCCAGAACGGCACG GTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGCCATGCTCACGGTGTTCCAGTGCATC ACCATGGAGGGCTGGACGGACGTGCTGTACTGGATGCAGGACGCTATGGGCTATGAGTTACCCTGGGTGTATTTTGTCAGTCTGGTCATC TTTGGATCCTTTTTCGTTCTAAATCTGGTTCTCGGTGTGTTGAGCGGAGAGTTTTCCAAAGAGAGGGAGAAGGCCAAGGCCCGGGGAGAT TTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCACTCAGGCCGAAGACATCGATCCTGAG AATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACAAGTGAATCACCCGGCAGTTCTGAGG CGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAAAGCTGTTACCTTTATAGATCTTACT ACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAATCCGGGAAGATCTCTTAAAAGCTTTA AATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGCTGTAAAAGCACTCAAGGCTGCAGGC TGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACGATTAGAAGAGATCAGATTGGCTGTG GAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGAAGGATCAGATTTTATTAAGACCTCT ACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAG ATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGG CTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCA GCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_90_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000406454_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWMQDAMGYELPWVYFVSLVIFGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- >12299_12299_91_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000480911_DERA_chr12_16109870_ENST00000428559_length(transcript)=2881nt_BP=1415nt CTCCTATTAAAACCATTTTTGGTCCATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAG CCCACGCCCCGCCCATGCCAACATGAATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTG GCAGGCGGCCATCGACGCAGCCCGGCAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAA GCGGCAGCAATATGGGAAACCCAAGAAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCC CATCCGGAGGGCCTGCATCAGCATTGTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGC GATCTATATTCCCTTTCCAGAAGATGATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGT GGAAGCGTTTTTAAAAGTAATCGCCTATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAAT TGTGGTTGTGGGGCTTTTTAGTGCAATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGA TGTGAAGGCGCTGAGGGCCTTCCGCGTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCAT CAAGGCCATGGTCCCCCTGCTGCACATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGG GAAGATGCACAAGACCTGCTACAACCAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCA CGGGCGGCAGTGCCAGAACGGCACGGTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGC CATGCTCACGGTGTTCCAGTGCATCACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTG GATCTATTTTGTTACACTAATCATCATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAG GGAGAAGGCCAAGGCCCGGGGAGATTTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCAC TCAGGCCGAAGACATCGATCCTGAGAATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACA AGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAA AGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAAT CCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGC TGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACG ATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGA AGCCCTGTATGATGAGATTCGTCAGTTTCGCAAGGCCTGTGGGGAGGCTCATCTTAAAACTATATTAGCGACAGGAGAACTTGGAACTCT TACTAATGTCTATAAAGCCAGTATGATAGCAATGATGGCAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCAC CTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCAT CCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGG TGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTA AATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATT GGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCAT TTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGT TAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAA CATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTG GATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTAAAATTTTTAAAATAATAA A >12299_12299_91_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000480911_DERA_chr12_16109870_ENST00000428559_length(amino acids)=771AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEALYDEIRQ FRKACGEAHLKTILATGELGTLTNVYKASMIAMMAGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLA WLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHDLPMS -------------------------------------------------------------- >12299_12299_92_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000480911_DERA_chr12_16109870_ENST00000532964_length(transcript)=2217nt_BP=1415nt CTCCTATTAAAACCATTTTTGGTCCATGGTCAATGAGAATACGAGGATGTACATTCCAGAGGAAAACCACCAAGGTTCCAACTATGGGAG CCCACGCCCCGCCCATGCCAACATGAATGCCAATGCGGCAGCGGGGCTGGCCCCTGAGCACATCCCCACCCCGGGGGCTGCCCTGTCGTG GCAGGCGGCCATCGACGCAGCCCGGCAGGCTAAGCTGATGGGCAGCGCTGGCAATGCGACCATCTCCACAGTCAGCTCCACGCAGCGGAA GCGGCAGCAATATGGGAAACCCAAGAAGCAGGGCAGCACCACGGCCACACGCCCGCCCCGAGCCCTGCTCTGCCTGACCCTGAAGAACCC CATCCGGAGGGCCTGCATCAGCATTGTCGAATGGAAACCATTTGAAATAATTATTTTACTGACTATTTTTGCCAATTGTGTGGCCTTAGC GATCTATATTCCCTTTCCAGAAGATGATTCCAACGCCACCAATTCCAACCTGGAACGAGTGGAATATCTCTTTCTCATAATTTTTACGGT GGAAGCGTTTTTAAAAGTAATCGCCTATGGACTCCTCTTTCACCCCAATGCCTACCTCCGCAACGGCTGGAACCTACTAGATTTTATAAT TGTGGTTGTGGGGCTTTTTAGTGCAATTTTAGAACAAGCAACCAAAGCAGATGGGGCAAACGCTCTCGGAGGGAAAGGGGCCGGATTTGA TGTGAAGGCGCTGAGGGCCTTCCGCGTGCTGCGCCCCCTGCGGCTGGTGTCCGGAGTCCCAAGTCTCCAGGTGGTCCTGAATTCCATCAT CAAGGCCATGGTCCCCCTGCTGCACATCGCCCTGCTTGTGCTGTTTGTCATCATCATCTACGCCATCATCGGCTTGGAGCTCTTCATGGG GAAGATGCACAAGACCTGCTACAACCAGGAGGGCATAGCAGATGTTCCAGCAGAAGATGACCCTTCCCCTTGTGCGCTGGAAACGGGCCA CGGGCGGCAGTGCCAGAACGGCACGGTGTGCAAGCCCGGCTGGGATGGTCCCAAGCACGGCATCACCAACTTTGACAACTTTGCCTTCGC CATGCTCACGGTGTTCCAGTGCATCACCATGGAGGGCTGGACGGACGTGCTGTACTGGGTCAATGATGCCGTAGGAAGGGACTGGCCCTG GATCTATTTTGTTACACTAATCATCATAGGGTCATTTTTTGTACTTAACTTGGTTCTCGGTGTGCTTAGCGGAGAGTTTTCCAAAGAGAG GGAGAAGGCCAAGGCCCGGGGAGATTTCCAGAAGCTGCGGGAGAAGCAGCAGCTAGAAGAGGATCTCAAAGGCTACCTGGATTGGATCAC TCAGGCCGAAGACATCGATCCTGAGAATGAGGACGAAGGCATGGATGAGGAGAAGCCCCGAAACAACCTTAGCTGGATCTCCAAAATACA AGTGAATCACCCGGCAGTTCTGAGGCGTGCGGAACAAATCCAGGCTCGCAGAACCGTGAAAAAGGAGTGGCAGGCTGCTTGGCTCCTGAA AGCTGTTACCTTTATAGATCTTACTACACTTTCAGGTGATGATACATCTTCCAACATTCAAAGGCTCTGTTATAAAGCCAAATACCCAAT CCGGGAAGATCTCTTAAAAGCTTTAAATATGCATGATAAAGGCATTACTACAGCCGCCGTTTGTGTTTATCCCGCCCGGGTGTGTGATGC TGTAAAAGCACTCAAGGCTGCAGGCTGTAATATCCCTGTGGCATCAGTGGCCGCTGGATTTCCAGCTGGACAGACTCATTTGAAGACACG ATTAGAAGAGATCAGATTGGCTGTGGAAGATGGAGCTACAGAAATCGACGTGGTAATTAACAGAAGCTTGGTGCTGACAGGCCAGTGGGA AGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGA TTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGT AAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTA CCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAG >12299_12299_92_CACNA1C-DERA_CACNA1C_chr12_2622150_ENST00000480911_DERA_chr12_16109870_ENST00000532964_length(amino acids)=728AA_BP=464 MVNENTRMYIPEENHQGSNYGSPRPAHANMNANAAAGLAPEHIPTPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPK KQGSTTATRPPRALLCLTLKNPIRRACISIVEWKPFEIIILLTIFANCVALAIYIPFPEDDSNATNSNLERVEYLFLIIFTVEAFLKVIA YGLLFHPNAYLRNGWNLLDFIIVVVGLFSAILEQATKADGANALGGKGAGFDVKALRAFRVLRPLRLVSGVPSLQVVLNSIIKAMVPLLH IALLVLFVIIIYAIIGLELFMGKMHKTCYNQEGIADVPAEDDPSPCALETGHGRQCQNGTVCKPGWDGPKHGITNFDNFAFAMLTVFQCI TMEGWTDVLYWVNDAVGRDWPWIYFVTLIIIGSFFVLNLVLGVLSGEFSKEREKAKARGDFQKLREKQQLEEDLKGYLDWITQAEDIDPE NEDEGMDEEKPRNNLSWISKIQVNHPAVLRRAEQIQARRTVKKEWQAAWLLKAVTFIDLTTLSGDDTSSNIQRLCYKAKYPIREDLLKAL NMHDKGITTAAVCVYPARVCDAVKALKAAGCNIPVASVAAGFPAGQTHLKTRLEEIRLAVEDGATEIDVVINRSLVLTGQWEGSDFIKTS TGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYA AYHDLPMS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CACNA1C-DERA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000327702 | + | 6 | 48 | 829_876 | 305.3333333333333 | 2174.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000344100 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2180.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000347598 | + | 6 | 49 | 829_876 | 305.3333333333333 | 2187.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399591 | + | 6 | 46 | 829_876 | 305.3333333333333 | 2147.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399595 | + | 6 | 46 | 829_876 | 305.3333333333333 | 2147.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399597 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399601 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399603 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399606 | + | 6 | 48 | 829_876 | 305.3333333333333 | 2159.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399617 | + | 6 | 48 | 829_876 | 305.3333333333333 | 2174.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399621 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2158.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399629 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2156.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399637 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2158.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399638 | + | 6 | 48 | 829_876 | 305.3333333333333 | 2167.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399641 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399644 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399649 | + | 6 | 46 | 829_876 | 305.3333333333333 | 2145.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000399655 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2595428 | chr12:16109870 | ENST00000402845 | + | 6 | 47 | 829_876 | 305.3333333333333 | 2158.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000327702 | + | 9 | 48 | 829_876 | 463.3333333333333 | 2174.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000344100 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2180.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000347598 | + | 9 | 49 | 829_876 | 463.3333333333333 | 2187.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399591 | + | 9 | 46 | 829_876 | 463.3333333333333 | 2147.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399595 | + | 9 | 46 | 829_876 | 463.3333333333333 | 2147.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399597 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399601 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399603 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399606 | + | 9 | 48 | 829_876 | 463.3333333333333 | 2159.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399617 | + | 9 | 48 | 829_876 | 463.3333333333333 | 2174.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399621 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2158.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399629 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2156.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399637 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2158.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399638 | + | 9 | 48 | 829_876 | 463.3333333333333 | 2167.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399641 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399644 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399649 | + | 9 | 46 | 829_876 | 463.3333333333333 | 2145.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000399655 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2139.0 | STAC2 |
| Hgene | CACNA1C | chr12:2622150 | chr12:16109870 | ENST00000402845 | + | 9 | 47 | 829_876 | 463.3333333333333 | 2158.0 | STAC2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CACNA1C-DERA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CACNA1C-DERA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CACNA1C | C0005586 | Bipolar Disorder | 9 | CTD_human;PSYGENET |
| Hgene | CACNA1C | C1832916 | Timothy syndrome | 8 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | CACNA1C | C1142166 | Brugada Syndrome (disorder) | 7 | CLINGEN;GENOMICS_ENGLAND |
| Hgene | CACNA1C | C1269683 | Major Depressive Disorder | 6 | CTD_human;PSYGENET |
| Hgene | CACNA1C | C0011570 | Mental Depression | 5 | PSYGENET |
| Hgene | CACNA1C | C0011581 | Depressive disorder | 5 | PSYGENET |
| Hgene | CACNA1C | C0041696 | Unipolar Depression | 5 | PSYGENET |
| Hgene | CACNA1C | C0525045 | Mood Disorders | 5 | PSYGENET |
| Hgene | CACNA1C | C2678478 | Brugada Syndrome 3 | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | CACNA1C | C0005587 | Depression, Bipolar | 4 | CTD_human |
| Hgene | CACNA1C | C0024713 | Manic Disorder | 4 | CTD_human |
| Hgene | CACNA1C | C0036341 | Schizophrenia | 4 | PSYGENET |
| Hgene | CACNA1C | C0338831 | Manic | 4 | CTD_human |
| Hgene | CACNA1C | C0033975 | Psychotic Disorders | 3 | PSYGENET |
| Hgene | CACNA1C | C0178417 | Anhedonia | 2 | PSYGENET |
| Hgene | CACNA1C | C0002895 | Anemia, Sickle Cell | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0003257 | Antibody Deficiency Syndrome | 1 | CTD_human |
| Hgene | CACNA1C | C0003469 | Anxiety Disorders | 1 | CTD_human |
| Hgene | CACNA1C | C0003811 | Cardiac Arrhythmia | 1 | CTD_human |
| Hgene | CACNA1C | C0004352 | Autistic Disorder | 1 | CTD_human |
| Hgene | CACNA1C | C0007194 | Hypertrophic Cardiomyopathy | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0011574 | Involutional Depression | 1 | CTD_human |
| Hgene | CACNA1C | C0020538 | Hypertensive disease | 1 | CTD_human |
| Hgene | CACNA1C | C0020615 | Hypoglycemia | 1 | CTD_human |
| Hgene | CACNA1C | C0021051 | Immunologic Deficiency Syndromes | 1 | CTD_human |
| Hgene | CACNA1C | C0038441 | Stress Disorders, Traumatic | 1 | CTD_human |
| Hgene | CACNA1C | C0039070 | Syncope | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0085298 | Sudden Cardiac Death | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0151879 | Shortened QT interval | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0206762 | Limb Deformities, Congenital | 1 | CTD_human |
| Hgene | CACNA1C | C0234985 | Mental deterioration | 1 | CTD_human |
| Hgene | CACNA1C | C0271708 | Fasting Hypoglycemia | 1 | CTD_human |
| Hgene | CACNA1C | C0271710 | Reactive hypoglycemia | 1 | CTD_human |
| Hgene | CACNA1C | C0338656 | Impaired cognition | 1 | CTD_human |
| Hgene | CACNA1C | C0342788 | Renal carnitine transport defect | 1 | GENOMICS_ENGLAND |
| Hgene | CACNA1C | C0349204 | Nonorganic psychosis | 1 | PSYGENET |
| Hgene | CACNA1C | C0376280 | Anxiety States, Neurotic | 1 | CTD_human |
| Hgene | CACNA1C | C0392156 | Akathisia | 1 | CTD_human |
| Hgene | CACNA1C | C1270972 | Mild cognitive disorder | 1 | CTD_human |
| Hgene | CACNA1C | C1279420 | Anxiety neurosis (finding) | 1 | CTD_human |
| Hgene | CACNA1C | C1571983 | Involutional paraphrenia | 1 | CTD_human |
| Hgene | CACNA1C | C1571984 | Psychosis, Involutional | 1 | CTD_human |
| Hgene | CACNA1C | C3887612 | Psychomotor Agitation | 1 | CTD_human |
