
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:DDR1-ATP5J2-PTCD1 (FusionGDB2 ID:HG780TG100526740) |
Fusion Gene Summary for DDR1-ATP5J2-PTCD1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: DDR1-ATP5J2-PTCD1 | Fusion gene ID: hg780tg100526740 | Hgene | Tgene | Gene symbol | DDR1 | ATP5J2-PTCD1 | Gene ID | 780 | 100526740 |
| Gene name | discoidin domain receptor tyrosine kinase 1 | ATP5MF-PTCD1 readthrough | |
| Synonyms | CAK|CD167|DDR|EDDR1|HGK2|MCK10|NEP|NTRK4|PTK3|PTK3A|RTK6|TRKE | ATP5J2-PTCD1 | |
| Cytomap | ('DDR1')('ATP5J2-PTCD1') 6p21.33 | 7q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | epithelial discoidin domain-containing receptor 1CD167 antigen-like family member APTK3A protein tyrosine kinase 3Acell adhesion kinasemammary carcinoma kinase 10neuroepithelial tyrosine kinaseneurotrophic tyrosine kinase, receptor, type 4protein-t | ATP5J2-PTCD1 fusion proteinATP5J2-PTCD1 read-through transcriptATP5J2-PTCD1 readthrough | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000508472, ENST00000324771, ENST00000376567, ENST00000376568, ENST00000376569, ENST00000376570, ENST00000376575, ENST00000418800, ENST00000446312, ENST00000452441, ENST00000454612, ENST00000508312, ENST00000513240, ENST00000361741, ENST00000259875, ENST00000383377, ENST00000400410, ENST00000400411, ENST00000400414, ENST00000400486, ENST00000400488, ENST00000400489, ENST00000400491, ENST00000400492, ENST00000412329, ENST00000415092, ENST00000419412, ENST00000421229, ENST00000422628, ENST00000427053, ENST00000429699, ENST00000430933, ENST00000434428, ENST00000449518, ENST00000453510, ENST00000454774, ENST00000462241, ENST00000468225, ENST00000483193, ENST00000487780, ENST00000488414, ENST00000490712, ENST00000548133, ENST00000548693, ENST00000548962, ENST00000549026, ENST00000550384, ENST00000550395, ENST00000550666, ENST00000552068, ENST00000552434, ENST00000552721, ENST00000553015, | ||
| Fusion gene scores | * DoF score | 6 X 6 X 4=144 | 9 X 10 X 2=180 |
| # samples | 6 | 10 | |
| ** MAII score | log2(6/144*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/180*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: DDR1 [Title/Abstract] AND ATP5J2-PTCD1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | DDR1(30859822)-ATP5J2-PTCD1(99022679), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | DDR1-ATP5J2-PTCD1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. DDR1-ATP5J2-PTCD1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. DDR1-ATP5J2-PTCD1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. DDR1-ATP5J2-PTCD1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. DDR1-ATP5J2-PTCD1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | DDR1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 9659899|21044884 |
| Hgene | DDR1 | GO:0038083 | peptidyl-tyrosine autophosphorylation | 21044884 |
| Hgene | DDR1 | GO:0046777 | protein autophosphorylation | 9659899 |
 Fusion gene breakpoints across DDR1 (5'-gene) Fusion gene breakpoints across DDR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across ATP5J2-PTCD1 (3'-gene) Fusion gene breakpoints across ATP5J2-PTCD1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
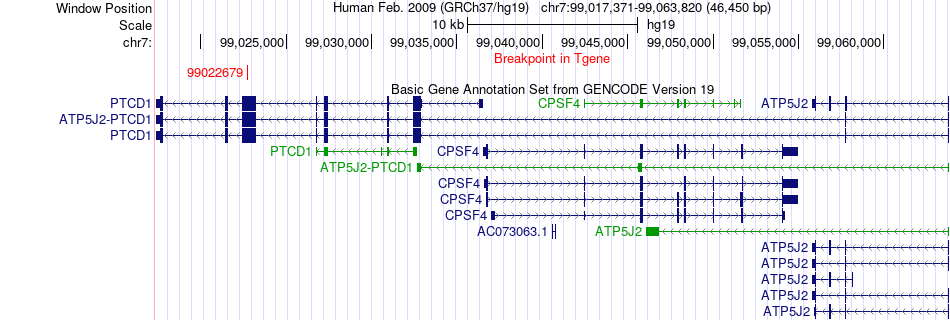 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
Top |
Fusion Gene ORF analysis for DDR1-ATP5J2-PTCD1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000324771 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.012783982 | 0.98721594 |
| ENST00000418800 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.012181659 | 0.98781836 |
| ENST00000454612 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.012021251 | 0.9879787 |
| ENST00000376569 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.011917049 | 0.98808295 |
| ENST00000376575 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.010613008 | 0.98938704 |
| ENST00000376570 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.010613008 | 0.98938704 |
| ENST00000376568 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.008711607 | 0.9912884 |
| ENST00000452441 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.007657961 | 0.99234205 |
| ENST00000508312 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.07044598 | 0.92955405 |
| ENST00000376567 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.009237992 | 0.99076205 |
| ENST00000513240 | ENST00000413834 | DDR1 | chr6 | 30859822 | + | ATP5J2-PTCD1 | chr7 | 99022679 | - | 0.007781317 | 0.99221873 |
Top |
Fusion Genomic Features for DDR1-ATP5J2-PTCD1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
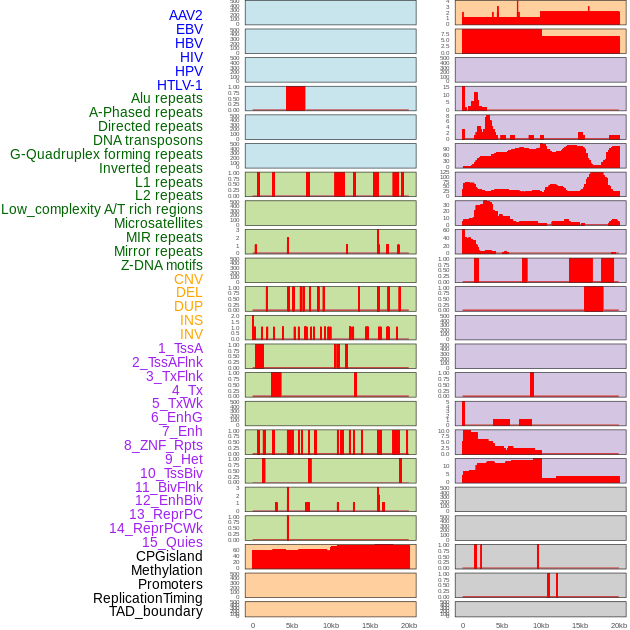 |
Top |
Fusion Protein Features for DDR1-ATP5J2-PTCD1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:30859822/chr7:99022679) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 377_415 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 476_601 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 377_415 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 476_601 | 0 | 914.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 377_415 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 476_601 | 0 | 877.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 377_415 | 0 | 895.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 476_601 | 0 | 895.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 377_415 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 476_601 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 377_415 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 476_601 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 377_415 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 476_601 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 377_415 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 476_601 | 0 | 646.3333333333334 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 377_415 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 476_601 | 0 | 920.0 | Compositional bias | Note=Gly/Pro-rich |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 610_905 | 0 | 646.3333333333334 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 610_905 | 0 | 914.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 610_905 | 0 | 877.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 610_905 | 0 | 895.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 610_905 | 0 | 646.3333333333334 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 610_905 | 0 | 646.3333333333334 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 610_905 | 0 | 646.3333333333334 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 610_905 | 0 | 646.3333333333334 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 610_905 | 0 | 920.0 | Domain | Protein kinase |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 481_484 | 0 | 646.3333333333334 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 481_484 | 0 | 914.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 481_484 | 0 | 877.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 481_484 | 0 | 895.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 481_484 | 0 | 646.3333333333334 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 481_484 | 0 | 646.3333333333334 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 481_484 | 0 | 646.3333333333334 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 481_484 | 0 | 646.3333333333334 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 481_484 | 0 | 920.0 | Motif | Note=PPxY motif |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 616_624 | 0 | 646.3333333333334 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 616_624 | 0 | 914.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 616_624 | 0 | 877.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 616_624 | 0 | 895.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 616_624 | 0 | 646.3333333333334 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 616_624 | 0 | 646.3333333333334 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 616_624 | 0 | 646.3333333333334 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 616_624 | 0 | 646.3333333333334 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 616_624 | 0 | 920.0 | Nucleotide binding | ATP |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 192_367 | 0 | 646.3333333333334 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 192_367 | 0 | 914.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 192_367 | 0 | 877.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 192_367 | 0 | 895.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 192_367 | 0 | 646.3333333333334 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 192_367 | 0 | 646.3333333333334 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 192_367 | 0 | 646.3333333333334 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 192_367 | 0 | 646.3333333333334 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 192_367 | 0 | 920.0 | Region | Note=DS-like domain |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 21_417 | 0 | 646.3333333333334 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 439_913 | 0 | 646.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 21_417 | 0 | 914.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 439_913 | 0 | 914.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 21_417 | 0 | 877.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 439_913 | 0 | 877.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 21_417 | 0 | 895.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 439_913 | 0 | 895.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 21_417 | 0 | 646.3333333333334 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 439_913 | 0 | 646.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 21_417 | 0 | 646.3333333333334 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 439_913 | 0 | 646.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 21_417 | 0 | 646.3333333333334 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 439_913 | 0 | 646.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 21_417 | 0 | 646.3333333333334 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 439_913 | 0 | 646.3333333333334 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 21_417 | 0 | 920.0 | Topological domain | Extracellular |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 439_913 | 0 | 920.0 | Topological domain | Cytoplasmic |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000259875 | + | 1 | 19 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000324771 | + | 1 | 20 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376567 | + | 1 | 16 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376568 | + | 1 | 18 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376569 | + | 1 | 18 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376570 | + | 1 | 19 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000376575 | + | 1 | 20 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000383377 | + | 1 | 20 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400410 | + | 1 | 16 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400411 | + | 1 | 17 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400414 | + | 1 | 19 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400486 | + | 1 | 16 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400488 | + | 1 | 17 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400489 | + | 1 | 19 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400491 | + | 1 | 19 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000400492 | + | 1 | 20 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000412329 | + | 1 | 20 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000415092 | + | 1 | 17 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000418800 | + | 1 | 17 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000419412 | + | 1 | 16 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000421229 | + | 1 | 19 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000427053 | + | 1 | 19 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000429699 | + | 1 | 19 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000430933 | + | 1 | 20 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000446312 | + | 1 | 18 | 418_438 | 0 | 646.3333333333334 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000449518 | + | 1 | 19 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000452441 | + | 1 | 19 | 418_438 | 0 | 914.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000453510 | + | 1 | 16 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454612 | + | 1 | 21 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000454774 | + | 1 | 17 | 418_438 | 0 | 877.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000508312 | + | 1 | 17 | 418_438 | 0 | 895.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000513240 | + | 1 | 17 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548133 | + | 1 | 20 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000548962 | + | 1 | 18 | 418_438 | 0 | 646.3333333333334 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000549026 | + | 1 | 18 | 418_438 | 0 | 646.3333333333334 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550384 | + | 1 | 18 | 418_438 | 0 | 646.3333333333334 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000550395 | + | 1 | 20 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552068 | + | 1 | 20 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000552721 | + | 1 | 18 | 418_438 | 0 | 646.3333333333334 | Transmembrane | Helical |
| Hgene | DDR1 | chr6:30859822 | chr7:99022679 | ENST00000553015 | + | 1 | 20 | 418_438 | 0 | 920.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for DDR1-ATP5J2-PTCD1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >21841_21841_1_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000324771_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=2202nt_BP=1356nt CCTTTGGTTCTGTTTCACAGTTCTCAGCCTCCCCTCCCTTTCTCCACAGCCAGGCTGCTCAGTCCCTCTCTGCGGGGGCCTAGAGGCTCG GTGAGGGGAGCGGGACTTGGTTGCCATGGTCACATTGAAGCCAGCCGCAGCTGGCCCGGGCAGCTGCTCCTCCTGGGCCCGGGGCCCCGG ACGCTCGGACAAAGCCAGGCAGCGTTGGCAGCCCCAGACCCGACCCCAAAGGCCTGAGACTGGGGTGACTGGGACCTAAGAGAATCCTGA GCTGGAGGCCCCCGACAGCTGCTCTCGGGAGCCGCCTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGC TCCCTCCGCCTCCCCCGCCCCTCGCCCCGCCGCCGAAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAG GTGTCTGAAGGTGGCTATTCACTGAGCGATGGGGTTGGACTTGAAGGAATGCCAAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGAT CAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATC CTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTG CCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGC AGGTGGATCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCT ACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTG AGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTC TGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCG TGTACCTCAACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGC TATGACTATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCT ATGCAGTACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAG GAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGG GGCAGGTGATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTG CCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCC CAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCA TCAACGCGGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGG TCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCC TTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGAC CTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGC CACAGGCCGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_1_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000324771_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_2_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376567_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1995nt_BP=1149nt GGCTTTAGGAAGTTATTTAACTGATCTCTGCCCTAGTTTCTTCATGTGTTAAATATGGATAGTAATAGTATCTACCTTATGAAGTGACTG TGAAGATAAAATTATGGATTCTGTTTAAGGGTTTAGGCCAGTGTCTGGCACAGGGGAAGCATTCTAAAAATATAGCTGATGCTGTTAAAC AATGACTGTTGTTGTTGTTTTACTGTTATTATCCCCAAAGCGGCCCATTCTGTCTGTTGCTGTCAGCTATGACTCAGTCCCCTGATTAAC TTACGCACCACCCATTTTATCCCCTGCAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGT CATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCA TGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCA GTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGG TGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTC GCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGC CCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCT GGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACG GACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCAC AGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTAC CTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTC CGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAAC AATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGA ACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCA CCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACT ACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCC AGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCAC CAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCG GTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTT CTCACAGACATGAAG >21841_21841_2_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376567_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=240AA_BP= MQRCCPHPLRPEGSGAMGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWC PAGSVFPKEEEYLQVDLQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLV RFYPRADRVMSVCLRVELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_3_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376568_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1949nt_BP=1103nt GGAAGGGAGGGAGCCGAGGCGAGGGGGAGGCGGCGCCTGGGCCCGAGCCGCCCCAGCCCTGGCTCCTCTCCCCGGAACAGGCCCCCGACA GCTGCTCTCGGGAGCCGCCTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCCGCCTCCCCCG CCCCTCGCCCCGCCGCCGAAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGAGATGCTGCCCCCACCC CCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACAT GAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTC CTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAA GGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAA GGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTC AGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCG GGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAAT GTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGC TGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGC TGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGA CAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATG ACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGA GCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCC CAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCA CATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGT CCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCG GGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGA AGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCA ACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_3_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376568_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_4_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376569_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1946nt_BP=1100nt AGAAGTAGGAGGGGCGTCTTCCCCTCGTGGGCCCTGAGCGGGACTGCAGCCAGCCCCCTGGGGCGCCAGCTTTGGAGGCCCCCGACAGCT GCTCTCGGGAGCCGCCTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCCGCCTCCCCCGCCC CTCGCCCCGCCGCCGAAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGAGATGCTGCCCCCACCCCCT TAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAA GGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTG GTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGA GGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGA GTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGG CAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGT CATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTA TTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGC GGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGA GGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAG TGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACG GATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCT GGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAA GGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACAT CTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCC GGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGA GTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGA GCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACC TGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_4_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376569_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_5_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376570_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1987nt_BP=1141nt GTCTTCCCCTCGTGGGCCCTGAGCGGGACTGCAGCCAGCCCCCTGGGGCGCCAGCTTTGGAGGCCCCCGACAGCTGCTCTCGGGAGCCGC CTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCCGCCTCCCCCGCCCCTCGCCCCGCCGCCG AAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGGTGTCTGAAGGTGGCTATTCACTGAGCGATGGGGT TGGACTTGAAGGAATGCCAAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTA CTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGAC CGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGG GATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTG GTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGG ATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATG GTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGAT GGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACC GTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTC CAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAA GATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAA GCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCT CGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGC AGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCG TGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTAT CTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCT CCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGA GGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGC AAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGA CATGAAG >21841_21841_5_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376570_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_6_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376575_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1987nt_BP=1141nt GTCTTCCCCTCGTGGGCCCTGAGCGGGACTGCAGCCAGCCCCCTGGGGCGCCAGCTTTGGAGGCCCCCGACAGCTGCTCTCGGGAGCCGC CTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCCGCCTCCCCCGCCCCTCGCCCCGCCGCCG AAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGGTGTCTGAAGGTGGCTATTCACTGAGCGATGGGGT TGGACTTGAAGGAATGCCAAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTA CTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGAC CGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGG GATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTG GTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGG ATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATG GTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGAT GGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACC GTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTC CAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAA GATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAA GCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCT CGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGC AGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCG TGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTAT CTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCT CCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGA GGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGC AAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGA CATGAAG >21841_21841_6_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000376575_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_7_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000418800_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=2067nt_BP=1221nt AGCCTAGAGCTGGGGCGAGGTGGGCATCACCACTAGGAATTTCTCCTGAGGCAGTGAGAAGAGGGGACAAAGGTTTCAGGACTCTCTAGC TCCTTCTGCTCTCCCCAGTGGACCCCTCTGTCTGGCACTGCCATGCCACTTAGCTGGGGTCAGCGTGGGCCTGGGGTGTGGAATGTCCCA CCAGGGTATGACGGGCTGTAGCTTGCCTGGCAGGCCTGTTGGGGCTTTCCCAGAGCACAGCTCCTGGAAGGAGGGGCTGTGGGCTGCCAG GTGAGGTGACTTGGGAAGCCTTGGCCCCACCCCCAGGCTGGCCCCACCCCCAGTCCAGCGTCTCCTGGGCCTAGATTCCCCAGCTGCTGT TCTCTGGAGGGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGC TCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCC CAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCT GGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCC AGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGA AGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGAC TGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGT CTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACCGTGGGCGGGG CTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTAT GTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGC TTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGG GACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAG TGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCAC AGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTC ACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCA TCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTG ACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTG ACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGC CTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_7_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000418800_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=244AA_BP= MLFSGGRCCPHPLRPEGSGAMGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGD GAWCPAGSVFPKEEEYLQVDLQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMV ARLVRFYPRADRVMSVCLRVELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_8_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000452441_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1913nt_BP=1067nt TGCTCTCGGGAGCCGCCTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCCGCCTCCCCCGCC CCTCGCCCCGCCGCCGAAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGGTGTCTGAAGGTGGCTATT CACTGAGCGATGGGGTTGGACTTGAAGGAATGCCAAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGA GGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGC CCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTT GGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACT GCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCG GGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGA CCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGG CTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCAC CTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGA GCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGA ACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGC AGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACA GCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGC CCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGC TCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAA GCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTT TGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGG ATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGC TGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTC TACAGCTTCTCACAGACATGAAG >21841_21841_8_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000452441_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_9_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000454612_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=2105nt_BP=1259nt AGATTCCCCAGCTGCTGTTCTCTGGAGGGGTTCCTGGGGCGGAGATGTGCAGTGGAGCGAGAACTTCCTGTGACCGTGACATTGTCTAGG TGGAACCGATCCCAGAAGTAGGAGGGGCGTCTTCCCCTCGTGGGCCCTGAGCGGGACTGCAGCCAGCCCCCTGGGGCGCCAGCTTTGGAG GCCCCCGACAGCTGCTCTCGGGAGCCGCCTCCCGACACCCGAGCCCCGCCGGCGCCTCCCGCTCCCGGCTCCCGGCTCCTGGCTCCCTCC GCCTCCCCCGCCCCTCGCCCCGCCGCCGAAGAGGCCCCGCTCCCGGGTCGGACGCCTGGGTCTGCCGGGAAGAGCGATGAGAGGTGTCTG AAGGTGGCTATTCACTGAGCGATGGGGTTGGACTTGAAGGAATGCCAAGAGATGCTGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGC TATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAA GTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCG CCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGA TCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCT GCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGT GGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGT AGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCT CAACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACT ATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGT ACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCC CGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGT GATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGG GAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGG TCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGC GGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCG CCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGC TGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGG GGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGC CGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_9_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000454612_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_10_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000508312_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1960nt_BP=1114nt CCCCTCTCACTCAGTGCGGAGGAACTGAGCCCCGGGAGGAGGTGCTCCTGTGCAGCCCCACTGAGTCAGCTCATCTATCGCCTGCCCTCC ACCTGGCCAGTCCCTGCGGGCATCTAACTGCTAAGCCTCCGCTCAGCCAACACCCAGTTGGTCAGTCTGGTCACAGTCCAGCAAAAAGAG GGACTGCCACTCTAACCCACCAGTGACACCACTCTTCCCGGCTGGATGGTCAATTAGCTCTGGCATGAGAGAATGTCACTGCCGAGATGC TGCCCCCACCCCCTTAGGCCCGAGGGATCAGGAGCTATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGA GATGCTGACATGAAGGGACATTTTGATCCTGCCAAGTGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCT GCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGCCACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCG GTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGATCTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGG GGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTGCGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAG GAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTGGTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCC CGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTAGAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTG GGGCAGACAATGTATTTATCTGAGGCCGTGTACCTCAACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAA GAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTATGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGA GTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTACCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAA GCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCCGCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAA GGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTGATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCC AGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGGAAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACA CACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGTCCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACC CCCAACACTCACATCTACAGTGCCCTCATCAACGCGGCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAG CAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGCCAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGT GGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCTGGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCT GGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGGGGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCA GACATTCTGCAACCTGGCCATCGGGTGCCACAGGCCGAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_10_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000508312_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=242AA_BP= MSLPRCCPHPLRPEGSGAMGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGA WCPAGSVFPKEEEYLQVDLQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVAR LVRFYPRADRVMSVCLRVELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- >21841_21841_11_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000513240_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(transcript)=1654nt_BP=808nt ATGGGACCAGAGGCCCTGTCATCTTTACTGCTGCTGCTCTTGGTGGCAAGTGGAGATGCTGACATGAAGGGACATTTTGATCCTGCCAAG TGCCGCTATGCCCTGGGCATGCAGGACCGGACCATCCCAGACAGTGACATCTCTGCTTCCAGCTCCTGGTCAGATTCCACTGCCGCCCGC CACAGCAGGTTGGAGAGCAGTGACGGGGATGGGGCCTGGTGCCCCGCAGGGTCGGTGTTTCCCAAGGAGGAGGAGTACTTGCAGGTGGAT CTACAACGACTGCACCTGGTGGCTCTGGTGGGCACCCAGGGACGGCATGCCGGGGGCCTGGGCAAGGAGTTCTCCCGGAGCTACCGGCTG CGTTACTCCCGGGATGGTCGCCGCTGGATGGGCTGGAAGGACCGCTGGGGTCAGGAGGTGATCTCAGGCAATGAGGACCCTGAGGGAGTG GTGCTGAAGGACCTTGGGCCCCCCATGGTTGCCCGACTGGTTCGCTTCTACCCCCGGGCTGACCGGGTCATGAGCGTCTGTCTGCGGGTA GAGCTCTATGGCTGCCTCTGGAGGGATGGACTCCTGTCTTACACCGCCCCTGTGGGGCAGACAATGTATTTATCTGAGGCCGTGTACCTC AACGACTCCACCTATGACGGACATACCGTGGGCGGGGCTGGATGACTTTAGGAAGAGTCAGGAGCTGCGGGTCTGGCCAGGCTATGACTA TGTGGGATGGAGCAACCACAGCTTCTCCAGTGGCTATGTGGAGATGGAGTTTGAGTTTGACCGGCTGAGGGCCTTCCAGGCTATGCAGTA CCAAGGGAAGAACACCTACCTGGAGAAGATTGACGGCTTCCGAGCCTATTACAAGCAGTGGCTGACAGTGATGCCCGCAGAGGAAACCCC GCACCCCTGGCAGAAGTTCCGGACCAAGCCCCAGGGGGACCAGGACACCGGCAAGGAGGCTGATGACGGATGTGCCCTTGGGGGCAGGTG ATGGGAGCACAGCTGGAACAATGTGCTCGGCCCCCAGTGCTCTGTGGGAGCCCCAGGACAAGTGAGCTGGTGTCACCTCCTGCCTGGGGG AAGAGCCAGGCCCTGAGGAACAGCCGCAGCGTGTCACAGGTGTTGGTGAGGACACACACTAGGCCCAAGGTGCCTGTGCTCCCAGCAGGT CCAAGTGCAGCTCCAGCCACCTTTGCGTGTCACCTTCACAAGTCCCAGGTGACCCCCAACACTCACATCTACAGTGCCCTCATCAACGCG GCCATCAGGAAGCTGAACTACACCTATCTCATCAGCATCTTGAAGGACATGAAGCAGAACAGGGTCCCGGTGAACGAAGTGGTCATCCGC CAGCTGGAGTTTGCAGCCCAGTACCCTCCCACCTTTGACCGGACTGGCCGAGGTGGTGGAGTCCGGGAGTCCTGCAGAGTCCTTGCTGCT GGCCCTCCTGGATGAGCACCAGGTAGAGGCCGACCTGACATTCTTTAACACGCTGGTGAGAAAGAAGAGCAAGCTGGGAGACCTGGAGGG GGCCAAGGCGCTGTTGCCGGTCCTGGCAAAGAGGGGCCTCGTCCCCAACCTGCAGACATTCTGCAACCTGGCCATCGGGTGCCACAGGCC GAAGGACGGTCTACAGCTTCTCACAGACATGAAG >21841_21841_11_DDR1-ATP5J2-PTCD1_DDR1_chr6_30859822_ENST00000513240_ATP5J2-PTCD1_chr7_99022679_ENST00000413834_length(amino acids)=224AA_BP= MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAARHSRLESSDGDGAWCPAGSVFPKEEEYLQVD LQRLHLVALVGTQGRHAGGLGKEFSRSYRLRYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGAG -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for DDR1-ATP5J2-PTCD1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for DDR1-ATP5J2-PTCD1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for DDR1-ATP5J2-PTCD1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | DDR1 | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Hgene | DDR1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Hgene | DDR1 | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| Hgene | DDR1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Hgene | DDR1 | C0036341 | Schizophrenia | 1 | PSYGENET |
| Hgene | DDR1 | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Hgene | DDR1 | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Hgene | DDR1 | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Hgene | DDR1 | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Hgene | DDR1 | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Hgene | DDR1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Hgene | DDR1 | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
