
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:COLEC11-SOX7 (FusionGDB2 ID:HG78989TG83595) |
Fusion Gene Summary for COLEC11-SOX7 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: COLEC11-SOX7 | Fusion gene ID: hg78989tg83595 | Hgene | Tgene | Gene symbol | COLEC11 | SOX7 | Gene ID | 78989 | 83595 |
| Gene name | collectin subfamily member 11 | SRY-box transcription factor 7 | |
| Synonyms | 3MC2|CL-K1-I|CL-K1-II|CL-K1-IIa|CL-K1-IIb|CLK1 | - | |
| Cytomap | ('COLEC11')('SOX7') 2p25.3 | 8p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | collectin-11Collectin K1collectin kidney protein 1collectin sub-family member 11 | transcription factor SOX-7SRY (sex determining region Y)-box 7SRY-box 7 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000487365, ENST00000236693, ENST00000349077, ENST00000382062, ENST00000418971, ENST00000402794, ENST00000402922, ENST00000403096, ENST00000404205, | ||
| Fusion gene scores | * DoF score | 3 X 2 X 2=12 | 5 X 5 X 3=75 |
| # samples | 2 | 5 | |
| ** MAII score | log2(2/12*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: COLEC11 [Title/Abstract] AND SOX7 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | COLEC11(3652060)-SOX7(10692285), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | COLEC11-SOX7 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COLEC11-SOX7 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. COLEC11-SOX7 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. COLEC11-SOX7 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. COLEC11-SOX7 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. COLEC11-SOX7 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | COLEC11 | GO:0006956 | complement activation | 23954398 |
| Hgene | COLEC11 | GO:0019730 | antimicrobial humoral response | 20956340 |
| Tgene | SOX7 | GO:0001706 | endoderm formation | 18682240 |
| Tgene | SOX7 | GO:0008285 | negative regulation of cell proliferation | 19108950 |
| Tgene | SOX7 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | 19108950 |
| Tgene | SOX7 | GO:0045892 | negative regulation of transcription, DNA-templated | 19108950 |
| Tgene | SOX7 | GO:0060828 | regulation of canonical Wnt signaling pathway | 19108950 |
 Fusion gene breakpoints across COLEC11 (5'-gene) Fusion gene breakpoints across COLEC11 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
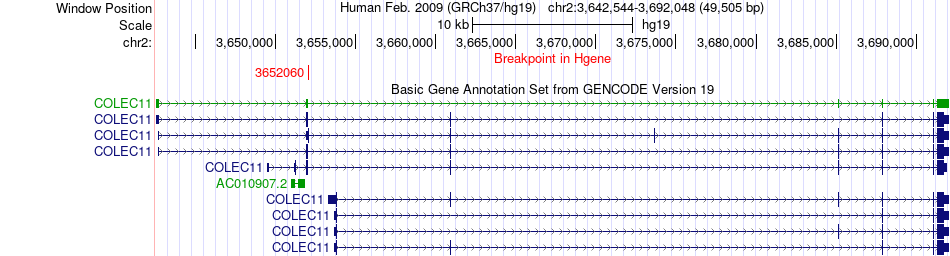 |
 Fusion gene breakpoints across SOX7 (3'-gene) Fusion gene breakpoints across SOX7 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
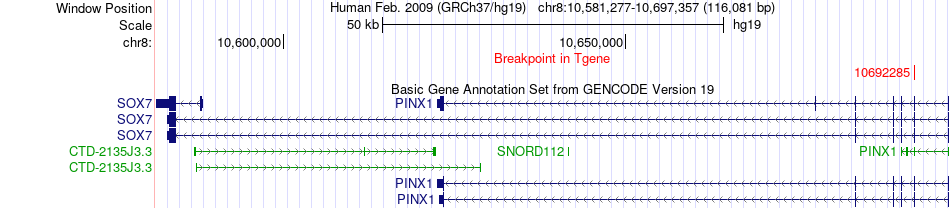 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-X6-A8C6-01A | COLEC11 | chr2 | 3652060 | - | SOX7 | chr8 | 10692285 | - |
| ChimerDB4 | SARC | TCGA-X6-A8C6-01A | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
Top |
Fusion Gene ORF analysis for COLEC11-SOX7 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000487365 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 3UTR-3CDS | ENST00000487365 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 3UTR-intron | ENST00000487365 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 5CDS-intron | ENST00000236693 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 5CDS-intron | ENST00000349077 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 5CDS-intron | ENST00000382062 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| 5CDS-intron | ENST00000418971 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| Frame-shift | ENST00000236693 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| Frame-shift | ENST00000236693 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000349077 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000349077 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000382062 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000382062 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000418971 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| In-frame | ENST00000418971 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000402794 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000402794 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000402922 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000402922 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000403096 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000403096 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000404205 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-3CDS | ENST00000404205 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-intron | ENST00000402794 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-intron | ENST00000402922 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-intron | ENST00000403096 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
| intron-intron | ENST00000404205 | ENST00000304501 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000382062 | COLEC11 | chr2 | 3652060 | + | ENST00000554914 | SOX7 | chr8 | 10692285 | - | 2013 | 370 | 225 | 1673 | 482 |
| ENST00000382062 | COLEC11 | chr2 | 3652060 | + | ENST00000553390 | SOX7 | chr8 | 10692285 | - | 2013 | 370 | 225 | 1673 | 482 |
| ENST00000349077 | COLEC11 | chr2 | 3652060 | + | ENST00000554914 | SOX7 | chr8 | 10692285 | - | 1876 | 233 | 88 | 1536 | 482 |
| ENST00000349077 | COLEC11 | chr2 | 3652060 | + | ENST00000553390 | SOX7 | chr8 | 10692285 | - | 1876 | 233 | 88 | 1536 | 482 |
| ENST00000418971 | COLEC11 | chr2 | 3652060 | + | ENST00000554914 | SOX7 | chr8 | 10692285 | - | 2026 | 383 | 211 | 1686 | 491 |
| ENST00000418971 | COLEC11 | chr2 | 3652060 | + | ENST00000553390 | SOX7 | chr8 | 10692285 | - | 2026 | 383 | 211 | 1686 | 491 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000382062 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.019419411 | 0.98058057 |
| ENST00000382062 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.019419411 | 0.98058057 |
| ENST00000349077 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.017893447 | 0.98210657 |
| ENST00000349077 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.017893447 | 0.98210657 |
| ENST00000418971 | ENST00000554914 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.009311847 | 0.9906881 |
| ENST00000418971 | ENST00000553390 | COLEC11 | chr2 | 3652060 | + | SOX7 | chr8 | 10692285 | - | 0.009311847 | 0.9906881 |
Top |
Fusion Genomic Features for COLEC11-SOX7 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
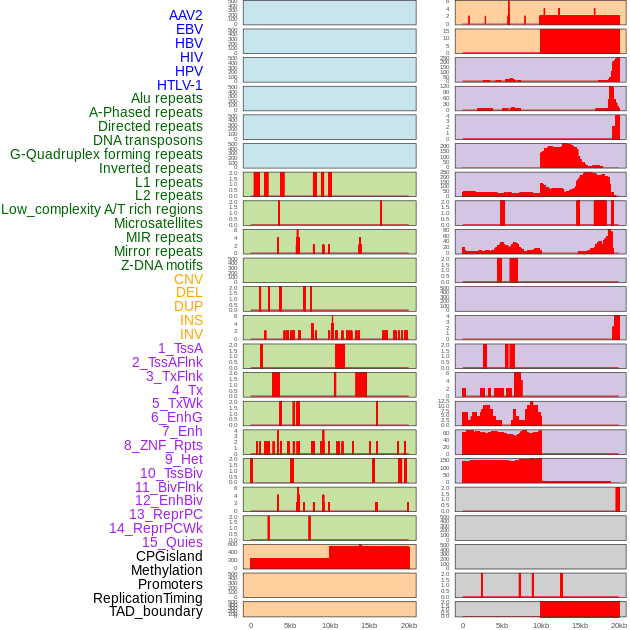 |
Top |
Fusion Protein Features for COLEC11-SOX7 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:3652060/chr8:10692285) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SOX7 | chr2:3652060 | chr8:10692285 | ENST00000304501 | 0 | 2 | 45_113 | 0 | 389.0 | DNA binding | HMG box | |
| Tgene | SOX7 | chr2:3652060 | chr8:10692285 | ENST00000304501 | 0 | 2 | 268_388 | 0 | 389.0 | Domain | Sox C-terminal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000236693 | + | 2 | 8 | 114_148 | 14 | 269.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000349077 | + | 2 | 7 | 114_148 | 43 | 272.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000382062 | + | 2 | 6 | 114_148 | 43 | 248.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402794 | + | 1 | 5 | 114_148 | 0 | 222.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402922 | + | 1 | 5 | 114_148 | 0 | 222.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000403096 | + | 1 | 6 | 114_148 | 0 | 246.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000404205 | + | 1 | 4 | 114_148 | 0 | 198.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000418971 | + | 3 | 8 | 114_148 | 57 | 286.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000236693 | + | 2 | 8 | 149_265 | 14 | 269.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000236693 | + | 2 | 8 | 65_112 | 14 | 269.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000349077 | + | 2 | 7 | 149_265 | 43 | 272.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000349077 | + | 2 | 7 | 65_112 | 43 | 272.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000382062 | + | 2 | 6 | 149_265 | 43 | 248.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000382062 | + | 2 | 6 | 65_112 | 43 | 248.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402794 | + | 1 | 5 | 149_265 | 0 | 222.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402794 | + | 1 | 5 | 65_112 | 0 | 222.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402922 | + | 1 | 5 | 149_265 | 0 | 222.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402922 | + | 1 | 5 | 65_112 | 0 | 222.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000403096 | + | 1 | 6 | 149_265 | 0 | 246.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000403096 | + | 1 | 6 | 65_112 | 0 | 246.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000404205 | + | 1 | 4 | 149_265 | 0 | 198.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000404205 | + | 1 | 4 | 65_112 | 0 | 198.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000418971 | + | 3 | 8 | 149_265 | 57 | 286.0 | Domain | C-type lectin |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000418971 | + | 3 | 8 | 65_112 | 57 | 286.0 | Domain | Note=Collagen-like |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000236693 | + | 2 | 8 | 252_254 | 14 | 269.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000349077 | + | 2 | 7 | 252_254 | 43 | 272.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000382062 | + | 2 | 6 | 252_254 | 43 | 248.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402794 | + | 1 | 5 | 252_254 | 0 | 222.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000402922 | + | 1 | 5 | 252_254 | 0 | 222.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000403096 | + | 1 | 6 | 252_254 | 0 | 246.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000404205 | + | 1 | 4 | 252_254 | 0 | 198.0 | Region | Carbohydrate binding |
| Hgene | COLEC11 | chr2:3652060 | chr8:10692285 | ENST00000418971 | + | 3 | 8 | 252_254 | 57 | 286.0 | Region | Carbohydrate binding |
Top |
Fusion Gene Sequence for COLEC11-SOX7 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >18483_18483_1_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000349077_SOX7_chr8_10692285_ENST00000553390_length(transcript)=1876nt_BP=233nt GCCATGAGGCGCCTGGGGGCAGTGTCCTCGCGGGCCAGCGACGGGCAGGACGCCCCGTTCGCCTAGCGCGTGCTCAGGAGTTGGTGTCCT GCCTGCGCTCAGGATGAGGGGGAATCTGGCCCTGGTGGGCGTTCTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCA GCCGGCTGGCGATGACGCCTGCTCTGTGCAGATCCTCGTCCCTGGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAA CACTGCCTGGAGTAATGACGATTCCAAGTTTGGCCAGCGGATGCTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGA GCAAGGAGCCACAGATCATATTAAAGTTCAAGTGAAAAATAACCACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGC CCATCAGGATGATTTTAACCAGCTTCTGGCCGAACTGAACACTTGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAA ATCTTTTAGCCTTGAGGAAAAGTCCAAAATCTCCAAAAACCGTGTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGAC GCTGTCCCAGAAGAGGCCGTACGTGGACGAGGCGGAGCGGCTGCGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCG CAGGAAGAAGCAGGCCAAGCGGCTGTGCAAGCGCGTGGACCCGGGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGA GAAGAGAAGCGGCAGCCGGGGGGCGCTGGGGGAGAAGGAGGACAGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTG CTACCACGAGGGGCCGGCTGGTGGTGGCGGCGGCGGCACCCCGAGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAAT GTCTCCCCTGGACGTGCTGGAGCCGGAGCAGACCTTCTTCTCCTCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCT GCCAGGGCACCCGTACTCACCGGAGTACGCCCCAAGCCCTCTCCACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGG CGTCTCCATGATGTCCCCTGTACCCGGCTGTCCCCCATCTCCTGCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCA AGCCCACCTGGGCCAGCTTTCCCCGCCTCCTGAGCACCCTGGCTTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACAT GGATCGCAATGAATTCGACCAGTATTTGAACACTCCTGGCCACCCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGT CTCCCAGGTGACACCAACGGGTCCCACAGAGACCAGCCTCATCTCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGT GTCATAGAGCTGGAGGCGCCCCGTCCGGTCAGCCCTCGCGCCCTCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCA GCTTTCCTCCCACCGCTCAGGGCAGGGAGGTCTGAACTGCGGCCCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCC TCTATCCCCTTCCCCACGTTCCAGCCCCTGCAGCCCACATTTTAAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCT GTCTCCCAGTGGAATGTTCACTGACGTCTTTTCTTGGTAGCCATCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_1_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000349077_SOX7_chr8_10692285_ENST00000553390_length(amino acids)=482AA_BP=48 MPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGWSKGKGLGAQ EQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKFTKGKSWKAL TLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSPGTALPSLRG CYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPLGSLALGQSP GVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATGAMALSGHVP VSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- >18483_18483_2_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000349077_SOX7_chr8_10692285_ENST00000554914_length(transcript)=1876nt_BP=233nt GCCATGAGGCGCCTGGGGGCAGTGTCCTCGCGGGCCAGCGACGGGCAGGACGCCCCGTTCGCCTAGCGCGTGCTCAGGAGTTGGTGTCCT GCCTGCGCTCAGGATGAGGGGGAATCTGGCCCTGGTGGGCGTTCTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCA GCCGGCTGGCGATGACGCCTGCTCTGTGCAGATCCTCGTCCCTGGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAA CACTGCCTGGAGTAATGACGATTCCAAGTTTGGCCAGCGGATGCTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGA GCAAGGAGCCACAGATCATATTAAAGTTCAAGTGAAAAATAACCACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGC CCATCAGGATGATTTTAACCAGCTTCTGGCCGAACTGAACACTTGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAA ATCTTTTAGCCTTGAGGAAAAGTCCAAAATCTCCAAAAACCGTGTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGAC GCTGTCCCAGAAGAGGCCGTACGTGGACGAGGCGGAGCGGCTGCGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCG CAGGAAGAAGCAGGCCAAGCGGCTGTGCAAGCGCGTGGACCCGGGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGA GAAGAGAAGCGGCAGCCGGGGGGCGCTGGGGGAGAAGGAGGACAGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTG CTACCACGAGGGGCCGGCTGGTGGTGGCGGCGGCGGCACCCCGAGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAAT GTCTCCCCTGGACGTGCTGGAGCCGGAGCAGACCTTCTTCTCCTCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCT GCCAGGGCACCCGTACTCACCGGAGTACGCCCCAAGCCCTCTCCACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGG CGTCTCCATGATGTCCCCTGTACCCGGCTGTCCCCCATCTCCTGCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCA AGCCCACCTGGGCCAGCTTTCCCCGCCTCCTGAGCACCCTGGCTTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACAT GGATCGCAATGAATTCGACCAGTATTTGAACACTCCTGGCCACCCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGT CTCCCAGGTGACACCAACGGGTCCCACAGAGACCAGCCTCATCTCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGT GTCATAGAGCTGGAGGCGCCCCGTCCGGTCAGCCCTCGCGCCCTCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCA GCTTTCCTCCCACCGCTCAGGGCAGGGAGGTCTGAACTGCGGCCCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCC TCTATCCCCTTCCCCACGTTCCAGCCCCTGCAGCCCACATTTTAAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCT GTCTCCCAGTGGAATGTTCACTGACGTCTTTTCTTGGTAGCCATCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_2_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000349077_SOX7_chr8_10692285_ENST00000554914_length(amino acids)=482AA_BP=48 MPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGWSKGKGLGAQ EQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKFTKGKSWKAL TLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSPGTALPSLRG CYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPLGSLALGQSP GVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATGAMALSGHVP VSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- >18483_18483_3_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000382062_SOX7_chr8_10692285_ENST00000553390_length(transcript)=2013nt_BP=370nt GGACGGTGGACGCAGCGCAGACAGGAAGCTCCCCGAGATAACGCTGCGGCCGGGCGGCCTGATTTGCTGGGCTGTCTGATGGCCCGGGCC GAGGCTTCTCCCTGCGCCTGGGACTGCGGCCGCCTCTCTAAATAGCAGCCATGAGGCGCCTGGGGGCAGTGTCCTCGCGGGCCAGCGACG GGCAGGACGCCCCGTTCGCCTAGCGCGTGCTCAGGAGTTGGTGTCCTGCCTGCGCTCAGGATGAGGGGGAATCTGGCCCTGGTGGGCGTT CTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCAGCCGGCTGGCGATGACGCCTGCTCTGTGCAGATCCTCGTCCCT GGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAACACTGCCTGGAGTAATGACGATTCCAAGTTTGGCCAGCGGATG CTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGAGCAAGGAGCCACAGATCATATTAAAGTTCAAGTGAAAAATAAC CACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGCCCATCAGGATGATTTTAACCAGCTTCTGGCCGAACTGAACACT TGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAAATCTTTTAGCCTTGAGGAAAAGTCCAAAATCTCCAAAAACCGT GTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGACGCTGTCCCAGAAGAGGCCGTACGTGGACGAGGCGGAGCGGCTG CGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCGCAGGAAGAAGCAGGCCAAGCGGCTGTGCAAGCGCGTGGACCCG GGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGAGAAGAGAAGCGGCAGCCGGGGGGCGCTGGGGGAGAAGGAGGAC AGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTGCTACCACGAGGGGCCGGCTGGTGGTGGCGGCGGCGGCACCCCG AGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAATGTCTCCCCTGGACGTGCTGGAGCCGGAGCAGACCTTCTTCTCC TCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCTGCCAGGGCACCCGTACTCACCGGAGTACGCCCCAAGCCCTCTC CACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGGCGTCTCCATGATGTCCCCTGTACCCGGCTGTCCCCCATCTCCT GCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCAAGCCCACCTGGGCCAGCTTTCCCCGCCTCCTGAGCACCCTGGC TTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACATGGATCGCAATGAATTCGACCAGTATTTGAACACTCCTGGCCAC CCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGTCTCCCAGGTGACACCAACGGGTCCCACAGAGACCAGCCTCATC TCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGTGTCATAGAGCTGGAGGCGCCCCGTCCGGTCAGCCCTCGCGCCC TCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCAGCTTTCCTCCCACCGCTCAGGGCAGGGAGGTCTGAACTGCGGC CCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCCTCTATCCCCTTCCCCACGTTCCAGCCCCTGCAGCCCACATTTT AAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCTGTCTCCCAGTGGAATGTTCACTGACGTCTTTTCTTGGTAGCCA TCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_3_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000382062_SOX7_chr8_10692285_ENST00000553390_length(amino acids)=482AA_BP=48 MPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGWSKGKGLGAQ EQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKFTKGKSWKAL TLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSPGTALPSLRG CYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPLGSLALGQSP GVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATGAMALSGHVP VSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- >18483_18483_4_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000382062_SOX7_chr8_10692285_ENST00000554914_length(transcript)=2013nt_BP=370nt GGACGGTGGACGCAGCGCAGACAGGAAGCTCCCCGAGATAACGCTGCGGCCGGGCGGCCTGATTTGCTGGGCTGTCTGATGGCCCGGGCC GAGGCTTCTCCCTGCGCCTGGGACTGCGGCCGCCTCTCTAAATAGCAGCCATGAGGCGCCTGGGGGCAGTGTCCTCGCGGGCCAGCGACG GGCAGGACGCCCCGTTCGCCTAGCGCGTGCTCAGGAGTTGGTGTCCTGCCTGCGCTCAGGATGAGGGGGAATCTGGCCCTGGTGGGCGTT CTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCAGCCGGCTGGCGATGACGCCTGCTCTGTGCAGATCCTCGTCCCT GGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAACACTGCCTGGAGTAATGACGATTCCAAGTTTGGCCAGCGGATG CTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGAGCAAGGAGCCACAGATCATATTAAAGTTCAAGTGAAAAATAAC CACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGCCCATCAGGATGATTTTAACCAGCTTCTGGCCGAACTGAACACT TGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAAATCTTTTAGCCTTGAGGAAAAGTCCAAAATCTCCAAAAACCGT GTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGACGCTGTCCCAGAAGAGGCCGTACGTGGACGAGGCGGAGCGGCTG CGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCGCAGGAAGAAGCAGGCCAAGCGGCTGTGCAAGCGCGTGGACCCG GGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGAGAAGAGAAGCGGCAGCCGGGGGGCGCTGGGGGAGAAGGAGGAC AGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTGCTACCACGAGGGGCCGGCTGGTGGTGGCGGCGGCGGCACCCCG AGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAATGTCTCCCCTGGACGTGCTGGAGCCGGAGCAGACCTTCTTCTCC TCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCTGCCAGGGCACCCGTACTCACCGGAGTACGCCCCAAGCCCTCTC CACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGGCGTCTCCATGATGTCCCCTGTACCCGGCTGTCCCCCATCTCCT GCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCAAGCCCACCTGGGCCAGCTTTCCCCGCCTCCTGAGCACCCTGGC TTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACATGGATCGCAATGAATTCGACCAGTATTTGAACACTCCTGGCCAC CCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGTCTCCCAGGTGACACCAACGGGTCCCACAGAGACCAGCCTCATC TCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGTGTCATAGAGCTGGAGGCGCCCCGTCCGGTCAGCCCTCGCGCCC TCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCAGCTTTCCTCCCACCGCTCAGGGCAGGGAGGTCTGAACTGCGGC CCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCCTCTATCCCCTTCCCCACGTTCCAGCCCCTGCAGCCCACATTTT AAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCTGTCTCCCAGTGGAATGTTCACTGACGTCTTTTCTTGGTAGCCA TCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_4_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000382062_SOX7_chr8_10692285_ENST00000554914_length(amino acids)=482AA_BP=48 MPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGWSKGKGLGAQ EQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKFTKGKSWKAL TLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSPGTALPSLRG CYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPLGSLALGQSP GVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATGAMALSGHVP VSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- >18483_18483_5_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000418971_SOX7_chr8_10692285_ENST00000553390_length(transcript)=2026nt_BP=383nt AGATTTGGGAGAAGGATTTCAAATGACTTCCCTTCGAGCTGCAGGGCTATCTTGGGCATCTTGACCTCCACACATTTCTGGGAGAACTCC TCGTGCAGCAGGACATCAGGTGGCAAAGGTGATCTGCCCACCTCAGCCTCCCAAAGTGCTGGAATTACAGGTGTGAGCCACTGCGCCTGG TCTTGTTTTCCAAGTTGTAACGCCCTTCACGATGAAGAAACAAAGAGGAGTTGGTGTCCTGCCTGCGCTCAGGATGAGGGGGAATCTGGC CCTGGTGGGCGTTCTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCAGCCGGCTGGCGATGACGCCTGCTCTGTGCA GATCCTCGTCCCTGGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAACACTGCCTGGAGTAATGACGATTCCAAGTT TGGCCAGCGGATGCTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGAGCAAGGAGCCACAGATCATATTAAAGTTCA AGTGAAAAATAACCACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGCCCATCAGGATGATTTTAACCAGCTTCTGGC CGAACTGAACACTTGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAAATCTTTTAGCCTTGAGGAAAAGTCCAAAAT CTCCAAAAACCGTGTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGACGCTGTCCCAGAAGAGGCCGTACGTGGACGA GGCGGAGCGGCTGCGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCGCAGGAAGAAGCAGGCCAAGCGGCTGTGCAA GCGCGTGGACCCGGGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGAGAAGAGAAGCGGCAGCCGGGGGGCGCTGGG GGAGAAGGAGGACAGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTGCTACCACGAGGGGCCGGCTGGTGGTGGCGG CGGCGGCACCCCGAGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAATGTCTCCCCTGGACGTGCTGGAGCCGGAGCA GACCTTCTTCTCCTCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCTGCCAGGGCACCCGTACTCACCGGAGTACGC CCCAAGCCCTCTCCACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGGCGTCTCCATGATGTCCCCTGTACCCGGCTG TCCCCCATCTCCTGCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCAAGCCCACCTGGGCCAGCTTTCCCCGCCTCC TGAGCACCCTGGCTTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACATGGATCGCAATGAATTCGACCAGTATTTGAA CACTCCTGGCCACCCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGTCTCCCAGGTGACACCAACGGGTCCCACAGA GACCAGCCTCATCTCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGTGTCATAGAGCTGGAGGCGCCCCGTCCGGTC AGCCCTCGCGCCCTCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCAGCTTTCCTCCCACCGCTCAGGGCAGGGAGG TCTGAACTGCGGCCCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCCTCTATCCCCTTCCCCACGTTCCAGCCCCTG CAGCCCACATTTTAAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCTGTCTCCCAGTGGAATGTTCACTGACGTCTT TTCTTGGTAGCCATCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_5_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000418971_SOX7_chr8_10692285_ENST00000553390_length(amino acids)=491AA_BP=57 MKKQRGVGVLPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGW SKGKGLGAQEQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKF TKGKSWKALTLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSP GTALPSLRGCYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPL GSLALGQSPGVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATG AMALSGHVPVSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- >18483_18483_6_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000418971_SOX7_chr8_10692285_ENST00000554914_length(transcript)=2026nt_BP=383nt AGATTTGGGAGAAGGATTTCAAATGACTTCCCTTCGAGCTGCAGGGCTATCTTGGGCATCTTGACCTCCACACATTTCTGGGAGAACTCC TCGTGCAGCAGGACATCAGGTGGCAAAGGTGATCTGCCCACCTCAGCCTCCCAAAGTGCTGGAATTACAGGTGTGAGCCACTGCGCCTGG TCTTGTTTTCCAAGTTGTAACGCCCTTCACGATGAAGAAACAAAGAGGAGTTGGTGTCCTGCCTGCGCTCAGGATGAGGGGGAATCTGGC CCTGGTGGGCGTTCTAATCAGCCTGGCCTTCCTGTCACTGCTGCCATCTGGACATCCTCAGCCGGCTGGCGATGACGCCTGCTCTGTGCA GATCCTCGTCCCTGGCCTCAAAGGTCGGCGGAAGCAGAAGTGGGCTGTGGATCCTCAGAACACTGCCTGGAGTAATGACGATTCCAAGTT TGGCCAGCGGATGCTAGAGAAGATGGGGTGGTCTAAAGGAAAGGGTTTAGGGGCTCAGGAGCAAGGAGCCACAGATCATATTAAAGTTCA AGTGAAAAATAACCACCTGGGACTCGGAGCTACCATCAATAATGAAGACAACTGGATTGCCCATCAGGATGATTTTAACCAGCTTCTGGC CGAACTGAACACTTGCCATGGGCAGGAAACCACAGATTCCTCGGACAAGAAGGAAAAGAAATCTTTTAGCCTTGAGGAAAAGTCCAAAAT CTCCAAAAACCGTGTTCACTATATGAAATTCACAAAAGGAAAGTCGTGGAAGGCGCTGACGCTGTCCCAGAAGAGGCCGTACGTGGACGA GGCGGAGCGGCTGCGCCTGCAGCACATGCAGGACTACCCCAACTACAAGTACCGGCCGCGCAGGAAGAAGCAGGCCAAGCGGCTGTGCAA GCGCGTGGACCCGGGCTTCCTTCTGAGCTCCCTCTCCCGGGACCAGAACGCCCTGCCGGAGAAGAGAAGCGGCAGCCGGGGGGCGCTGGG GGAGAAGGAGGACAGGGGTGAGTACTCCCCCGGCACTGCCCTGCCCAGCCTCCGGGGCTGCTACCACGAGGGGCCGGCTGGTGGTGGCGG CGGCGGCACCCCGAGCAGTGTGGACACGTACCCGTACGGGCTGCCCACACCTCCTGAAATGTCTCCCCTGGACGTGCTGGAGCCGGAGCA GACCTTCTTCTCCTCCCCCTGCCAGGAGGAGCATGGCCATCCCCGCCGCATCCCCCACCTGCCAGGGCACCCGTACTCACCGGAGTACGC CCCAAGCCCTCTCCACTGTAGCCACCCCCTGGGCTCCCTGGCCCTTGGCCAGTCCCCCGGCGTCTCCATGATGTCCCCTGTACCCGGCTG TCCCCCATCTCCTGCCTATTACTCCCCGGCCACCTACCACCCACTCCACTCCAACCTCCAAGCCCACCTGGGCCAGCTTTCCCCGCCTCC TGAGCACCCTGGCTTCGACGCCCTGGATCAACTGAGCCAGGTGGAACTCCTGGGGGACATGGATCGCAATGAATTCGACCAGTATTTGAA CACTCCTGGCCACCCAGACTCCGCCACAGGGGCCATGGCCCTCAGTGGGCATGTTCCGGTCTCCCAGGTGACACCAACGGGTCCCACAGA GACCAGCCTCATCTCCGTCCTGGCTGATGCCACGGCCACGTACTACAACAGCTACAGTGTGTCATAGAGCTGGAGGCGCCCCGTCCGGTC AGCCCTCGCGCCCTCTCCTTCTTGTGCCTTGAGTGGCAGAGGAGCCGTCCAGCCACACCAGCTTTCCTCCCACCGCTCAGGGCAGGGAGG TCTGAACTGCGGCCCCAGAGCCTTTGGCCTAAGCTGGACTCTCCTTATCCGAGTGCCGCCTCTATCCCCTTCCCCACGTTCCAGCCCCTG CAGCCCACATTTTAAGTATATTCCTTCAAGTGAGTTTTCCTCCAGCCCCTGAGAGTTGCTGTCTCCCAGTGGAATGTTCACTGACGTCTT TTCTTGGTAGCCATCATCGAAACTAATGGGGGGACAGACTTGATAG >18483_18483_6_COLEC11-SOX7_COLEC11_chr2_3652060_ENST00000418971_SOX7_chr8_10692285_ENST00000554914_length(amino acids)=491AA_BP=57 MKKQRGVGVLPALRMRGNLALVGVLISLAFLSLLPSGHPQPAGDDACSVQILVPGLKGRRKQKWAVDPQNTAWSNDDSKFGQRMLEKMGW SKGKGLGAQEQGATDHIKVQVKNNHLGLGATINNEDNWIAHQDDFNQLLAELNTCHGQETTDSSDKKEKKSFSLEEKSKISKNRVHYMKF TKGKSWKALTLSQKRPYVDEAERLRLQHMQDYPNYKYRPRRKKQAKRLCKRVDPGFLLSSLSRDQNALPEKRSGSRGALGEKEDRGEYSP GTALPSLRGCYHEGPAGGGGGGTPSSVDTYPYGLPTPPEMSPLDVLEPEQTFFSSPCQEEHGHPRRIPHLPGHPYSPEYAPSPLHCSHPL GSLALGQSPGVSMMSPVPGCPPSPAYYSPATYHPLHSNLQAHLGQLSPPPEHPGFDALDQLSQVELLGDMDRNEFDQYLNTPGHPDSATG AMALSGHVPVSQVTPTGPTETSLISVLADATATYYNSYSVS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for COLEC11-SOX7 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for COLEC11-SOX7 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for COLEC11-SOX7 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | COLEC11 | C0796279 | Carnevale syndrome | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | COLEC11 | C0022283 | Incontinentia Pigmenti Achromians | 1 | CTD_human |
| Hgene | COLEC11 | C0036305 | Schamberg Disease | 1 | CTD_human |
| Hgene | COLEC11 | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | COLEC11 | C0549567 | Pigmentation Disorders | 1 | CTD_human |
| Hgene | COLEC11 | C0796032 | Malpuech facial clefting syndrome | 1 | CTD_human |
| Hgene | COLEC11 | C0796059 | Oculopalatoskeletal syndrome | 1 | CTD_human |
| Hgene | COLEC11 | C4303860 | Craniofacial ulnar renal syndrome | 1 | ORPHANET |
