
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:EHMT1-SPATA17 (FusionGDB2 ID:HG79813TG128153) |
Fusion Gene Summary for EHMT1-SPATA17 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: EHMT1-SPATA17 | Fusion gene ID: hg79813tg128153 | Hgene | Tgene | Gene symbol | EHMT1 | SPATA17 | Gene ID | 79813 | 128153 |
| Gene name | euchromatic histone lysine methyltransferase 1 | spermatogenesis associated 17 | |
| Synonyms | EHMT1-IT1|EUHMTASE1|Eu-HMTase1|FP13812|GLP|GLP1|KLEFS1|KMT1D | CFAP305|FAP305|IQCH|MOT17|MSRG-11|MSRG11 | |
| Cytomap | ('EHMT1')('SPATA17') 9q34.3 | 1q41 | |
| Type of gene | protein-coding | protein-coding | |
| Description | histone-lysine N-methyltransferase EHMT1EHMT1 intronic transcript 1G9a-like protein 1H3-K9-HMTase 5euchromatic histone-lysine N-methyltransferase 1histone H3-K9 methyltransferase 5histone-lysine N-methyltransferase, H3 lysine-9 specific 5lysine N-m | spermatogenesis-associated protein 17IQ motif containing Hspermatogenesis-related protein 11 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000371394, ENST00000460843, ENST00000462484, ENST00000334856, | ||
| Fusion gene scores | * DoF score | 15 X 9 X 8=1080 | 7 X 9 X 6=378 |
| # samples | 20 | 7 | |
| ** MAII score | log2(20/1080*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/378*10)=-2.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: EHMT1 [Title/Abstract] AND SPATA17 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | EHMT1(140513501)-SPATA17(217856600), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | EHMT1-SPATA17 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EHMT1-SPATA17 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EHMT1-SPATA17 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. EHMT1-SPATA17 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EHMT1 | GO:0006325 | chromatin organization | 12004135 |
| Hgene | EHMT1 | GO:0016571 | histone methylation | 12004135 |
| Hgene | EHMT1 | GO:0018027 | peptidyl-lysine dimethylation | 20118233 |
 Fusion gene breakpoints across EHMT1 (5'-gene) Fusion gene breakpoints across EHMT1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
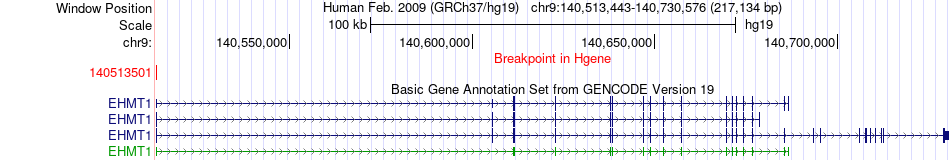 |
 Fusion gene breakpoints across SPATA17 (3'-gene) Fusion gene breakpoints across SPATA17 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
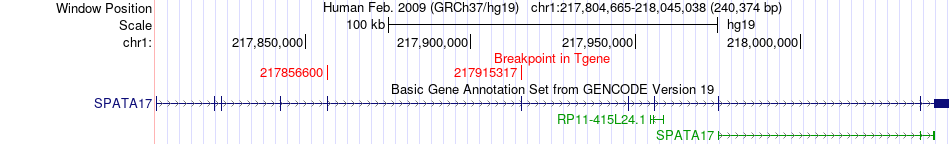 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-1924-01A | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| ChimerDB4 | OV | TCGA-24-1924-01A | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
Top |
Fusion Gene ORF analysis for EHMT1-SPATA17 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000371394 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 3UTR-3CDS | ENST00000371394 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| 3UTR-intron | ENST00000371394 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 3UTR-intron | ENST00000371394 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| 5CDS-intron | ENST00000460843 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 5CDS-intron | ENST00000460843 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| 5CDS-intron | ENST00000462484 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 5CDS-intron | ENST00000462484 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| 5UTR-3CDS | ENST00000334856 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 5UTR-3CDS | ENST00000334856 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| 5UTR-intron | ENST00000334856 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| 5UTR-intron | ENST00000334856 | ENST00000471021 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| Frame-shift | ENST00000460843 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| Frame-shift | ENST00000462484 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915317 | + |
| In-frame | ENST00000460843 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
| In-frame | ENST00000462484 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000462484 | EHMT1 | chr9 | 140513501 | + | ENST00000366933 | SPATA17 | chr1 | 217856600 | + | 5530 | 58 | 22 | 852 | 276 |
| ENST00000460843 | EHMT1 | chr9 | 140513501 | + | ENST00000366933 | SPATA17 | chr1 | 217856600 | + | 5520 | 48 | 12 | 842 | 276 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000462484 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + | 0.000715393 | 0.9992847 |
| ENST00000460843 | ENST00000366933 | EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856600 | + | 0.000716666 | 0.9992834 |
Top |
Fusion Genomic Features for EHMT1-SPATA17 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915316 | + | 2.52E-06 | 0.9999975 |
| EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856599 | + | 1.49E-08 | 1 |
| EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217915316 | + | 2.52E-06 | 0.9999975 |
| EHMT1 | chr9 | 140513501 | + | SPATA17 | chr1 | 217856599 | + | 1.49E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
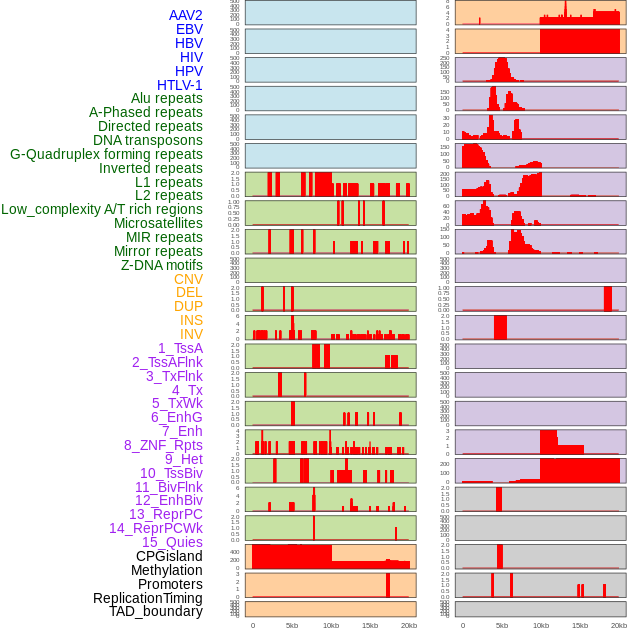 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
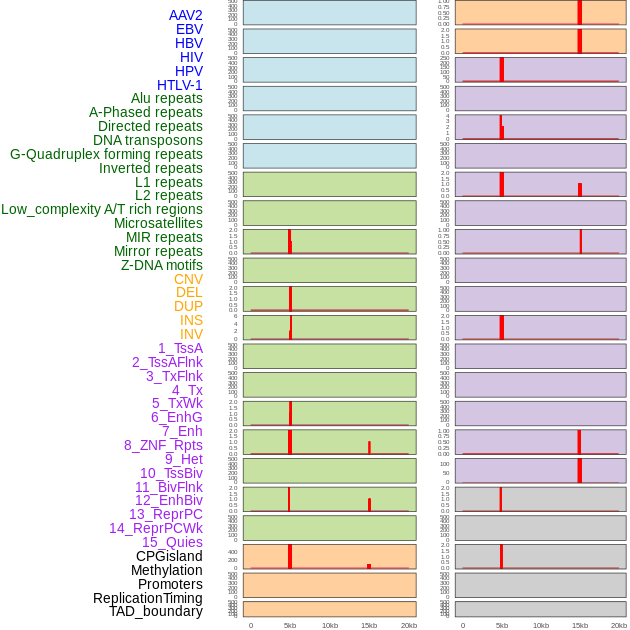 |
Top |
Fusion Protein Features for EHMT1-SPATA17 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:140513501/chr1:217856600) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1292_1295 | 0 | 826.0 | Compositional bias | Note=Poly-Ala |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 406_409 | 0 | 826.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 442_449 | 0 | 826.0 | Compositional bias | Note=Poly-Arg |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1292_1295 | 7 | 1299.0 | Compositional bias | Note=Poly-Ala |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 406_409 | 7 | 1299.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 442_449 | 7 | 1299.0 | Compositional bias | Note=Poly-Arg |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1292_1295 | 7 | 809.0 | Compositional bias | Note=Poly-Ala |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 406_409 | 7 | 809.0 | Compositional bias | Note=Poly-Glu |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 442_449 | 7 | 809.0 | Compositional bias | Note=Poly-Arg |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1060_1123 | 0 | 826.0 | Domain | Pre-SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1126_1243 | 0 | 826.0 | Domain | SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1060_1123 | 7 | 1299.0 | Domain | Pre-SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1126_1243 | 7 | 1299.0 | Domain | SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1060_1123 | 7 | 809.0 | Domain | Pre-SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1126_1243 | 7 | 809.0 | Domain | SET |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1136_1138 | 0 | 826.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1200_1201 | 0 | 826.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 905_907 | 0 | 826.0 | Region | Note=Histone H3K9me binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1136_1138 | 7 | 1299.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1200_1201 | 7 | 1299.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 905_907 | 7 | 1299.0 | Region | Note=Histone H3K9me binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1136_1138 | 7 | 809.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1200_1201 | 7 | 809.0 | Region | Note=S-adenosyl-L-methionine binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 905_907 | 7 | 809.0 | Region | Note=Histone H3K9me binding |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 737_766 | 0 | 826.0 | Repeat | Note=ANK 1 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 772_801 | 0 | 826.0 | Repeat | Note=ANK 2 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 805_834 | 0 | 826.0 | Repeat | Note=ANK 3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 838_868 | 0 | 826.0 | Repeat | Note=ANK 4 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 872_901 | 0 | 826.0 | Repeat | Note=ANK 5 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 905_934 | 0 | 826.0 | Repeat | Note=ANK 6 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 938_967 | 0 | 826.0 | Repeat | Note=ANK 7 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 971_1004 | 0 | 826.0 | Repeat | Note=ANK 8 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 737_766 | 7 | 1299.0 | Repeat | Note=ANK 1 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 772_801 | 7 | 1299.0 | Repeat | Note=ANK 2 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 805_834 | 7 | 1299.0 | Repeat | Note=ANK 3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 838_868 | 7 | 1299.0 | Repeat | Note=ANK 4 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 872_901 | 7 | 1299.0 | Repeat | Note=ANK 5 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 905_934 | 7 | 1299.0 | Repeat | Note=ANK 6 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 938_967 | 7 | 1299.0 | Repeat | Note=ANK 7 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 971_1004 | 7 | 1299.0 | Repeat | Note=ANK 8 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 737_766 | 7 | 809.0 | Repeat | Note=ANK 1 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 772_801 | 7 | 809.0 | Repeat | Note=ANK 2 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 805_834 | 7 | 809.0 | Repeat | Note=ANK 3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 838_868 | 7 | 809.0 | Repeat | Note=ANK 4 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 872_901 | 7 | 809.0 | Repeat | Note=ANK 5 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 905_934 | 7 | 809.0 | Repeat | Note=ANK 6 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 938_967 | 7 | 809.0 | Repeat | Note=ANK 7 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 971_1004 | 7 | 809.0 | Repeat | Note=ANK 8 |
| Tgene | SPATA17 | chr9:140513501 | chr1:217856600 | ENST00000366933 | 3 | 11 | 32_61 | 97 | 1879.3333333333333 | Domain | IQ 1 | |
| Tgene | SPATA17 | chr9:140513501 | chr1:217856600 | ENST00000366933 | 3 | 11 | 55_84 | 97 | 1879.3333333333333 | Domain | IQ 2 | |
| Tgene | SPATA17 | chr9:140513501 | chr1:217856600 | ENST00000366933 | 3 | 11 | 91_120 | 97 | 1879.3333333333333 | Domain | IQ 3 |
Top |
Fusion Gene Sequence for EHMT1-SPATA17 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >25589_25589_1_EHMT1-SPATA17_EHMT1_chr9_140513501_ENST00000460843_SPATA17_chr1_217856600_ENST00000366933_length(transcript)=5520nt_BP=48nt GGCGGGGCCACGCTGCGGGCCCGGGCCATGGCCGCCGCCGATGCCGAGATTCAGAGACGATGGCGAGGCTATAGGGTTCGGAAGTACCTC TTTAATTATTATTATTTGAAAGAGTACCTGAAAGTCGTTTCAGAGACCAATGATGCAATTAGGAAGGCACTGGAGGAGTTTGCAGAAATG AAAGAAAGAGAAGAGAAGAAGGCTAACCTCGAAAGGGAAGAGAAGAAAAGAGATTACCAAGCCCGAAAGATGCATTACCTCCTCAGCACA AAGCAGATTCCAGGAATATACAATTCACCCTTCAGAAAAGAGCCTGATCCATGGGAGCTGCAATTACAGAAGGCAAAGCCTTTAACACAC CGAAGACCTAAAGTTAAGCAGAAGGACTCCACCAGCCTTACTGATTGGCTAGCTTGTACAAGCGCCCGTTCTTTTCCTCGGTCTGAAATT CTACCACCTATTAATAGAAAGCAATGTCAGGGGCCCTTCCGAGATATCACCGAAGTATTAGAACAACGCTACAGGCCTTTGGAGCCAACG TTGCGGGTGGCAGAACCAATCGATGAGTTAAAGTTGGCCAGAGAGGAGCTCAGAAGAGAGGAATGGCTGCAAAATGTAAATGACAATATG TTTTTGCCATTTTCTTCATACCATAAAAATGAAAAGTACATCCCATCAATGCATTTATCAAGCAAGTATGGTCCTATTTCTTACAAAGAA CAATTCCGAAGTGAAAATCCTAAGAAATGGATCTGTGACAAGGATTTCCAGACTGTATTACCATCATTTGAGCTCTTCTCAAAGTATGGA AAATTATATTCAAAAGCTGGACAGATTGTATAAAGGCGTCAGAAGAAGAAACTGAAGCCATCTGCATTTTAAAACTTAACAGTTCTGAAA GGAAAACACAGATGAAGATCCTGTAGGAAATATACTTGCTATGATTCAATAAACTATAAAATTTTGTAGCTCTAGTCATGAATTAATTAT TGTGTGTATGTATTTGAAATCTCTTAGACATATCTTTTCAAACAAATACTTAAACTTGAGTCTGTTACCTCTGAGAATGTCTGTCACAAT CACAGCCCCTGGTGACCAACAGTGCCATAAATGGTTCCCCTGAAATACCCATATTCTGACCCCATCACCCGGGTCACAAATGATGACACC AGTGCTGGGTACCCAAATTAAGGGCGACTAACATGTAAGTAGGATGATGAATGGTTTGAAGTGTAAATTGTGTGAAATAGTTCAGCATGA AAGCTCTGCCCTGAGAAATAATGACTGATTATACCAGTTAGAAACCCTTCAGACTGTTGAACTTGAGACACACAGACACAACAGACACAG ACTAGTGGGTGTGGGAGCAGAAGCATAGGCACTCTCAAATAGAGAAGACATTTGCAAGTCTCTGCTGCCAAGGGAGCAAAGAGATAATCT CGATGCCAGGGCTGTGTCAGATCCTGCATGGCTCTGAACTACAGTTGGTTTCCAAAAAGCTGGGCTTTTGTGAGGTTTCTGTTTTTTATG CGATGCTTAGATTTTCCTGGAATCCTTGGAAGTTAACTCAACATTTGAGGTTCCTATATCCTTTAATCCCATCCCATGTGTATCTTTACA ATAAAACTTCCTGCTAGAGGTATCCTGAGGGAATCTTTGTTTCTTGCATCTGAAGAGAATAACACTAAACTTATTGCATATGATTTGGGG GTTGGATCCAATGATAGCACAGAGGGACAGGAAATGTGATTTATTGACCATAAAAGCTTTAATTTAAAATTGCTGGCCAGCGAAATCCAG ACTCTGGGAAATGTTATAGGCCAAATGATCTGTTTCTTCAACAAATAAATTACAAAACTGAGGGGAAAAAAAGGAACCTATTAAAAAGAC ATTATCTATCAATCACTGTGTGTGTACAGTTTTGAGCTCCTGATTTAAATAAACAAATTGTAAAATAAAAGATCTTTTGAAACAATTGGA AGTTTGAACACTGATTGGATATTGGTGGTATAAAAGAATTATTGGTTTTTTTTAGGTGTCATGGTATTGTTTTGGTTTACAAAGTTATAG ATATAAATGTTGAAATATTTACAGATGCAGTGATATGTTGGCTGGGATTTAATTTGAAAGTCAAAGAAGTGAGTTAAAATACATATACTC CTCCTCTTATAATCGGTTATGTGCCAATAAATCCATTGTAAATTGAAAATATCCTAAGTTGAAAATATTGTAAGTTAAAAATGCATTTAA TACAGCTAACTTACCAGACAGCTTAGCTTAGCCTCACCTACCTTAAATATGCTCAGAACACATACGTTAGCCTATAGTTGGGCAAAATCA TCTAACACAAAGCCTATTTTATAATATTGAACAGCCCATGTAATTTATTGAATATTGTACTAAAAGTGAAAAACTGAATAGTTGTATGCA TACTCAAAGTATGGTTTCTACTGAATGCATTGTAAACCAGAAAGTATCTGAGACAATGTTTTTTTTTTTTTTTTTTTTTTTTTTAATTTC TGAGACAGAGTCTCATTCTGTCACCCAGGTTGGAGTGCAGTGGCATGAACTCAGCTCACTGCAACCTTCACCTCCCAGGCTCAAGCAGTC CTCCCATCTCAGCCTCCCAAGTAGCTGGGACCACAGGCATGGGCCAACAAGCCTGGCTAAATATTTTGTACTTTTGGTAGACACGGGTTT TTACCATGTTGTGCAGGCTGGTCTTGAACGCCTGAGCTCAAGTGATCTGCCTGCCTCGGCCTCCCAAAGTACTGGGATTACAGGCATGAG TCACTGAACCCAGCCCTGAGACATGTCTCAATTAGTTTAGAAGTTTATTTGGCCAAGGTTAAGGACATGCCCTTGACACTGCACCTGTCA GTACAGGTCCTGACAACATGTGTCCAAGGTGGTCAAGCTATACAACTTGGTTTTATACATTTTAAGGAGACATAAGACATCAATAAATAC ATGGAAGATGTACATTGGTTCAGTCTGGAAAGGCAGGATGCCTCAAACTTGAAACAAGAGAATGGGGGGTGGGAGGATGTGGGAGATGCT TCCAGGTCATAGTTGGAGTCAAAGACTTTCCAATTGGCAATTGGTTCAAAGAATTTATCTAAAGACCTGGAATCCATAGAAGGCTTTACC TGGATTAAAATAAGGGGACCAAGGTTCTCATGCAGATGAAGCCTCCAGGTGGCAGACTTCAGAGAGCATAGATTGTAAATGTTTCTTATC TGACTTAAAAAGGTGTCAGACTCTTAGTAAATTATCTCCTGGATCAGTGAAAAGATGTGAAAAAAGGTTCTCTACAGAATGTAGATTTTC CCCATAAGAGATAGCTTCGCAGGGCCATTGCAAAATATGTCAAAAATATATATTTTAGGGTAAAATGCCTCAATTTATTTCAAGGCCTGA TATTTGTCATGTGATGCTATACTACAGTCAGGCTGAAACTTGGCATCTTATTGCTATGAAAAATCTTAAGATCTCTATTTTAATGTTAAT GCTGGTCACTGGTGCCTGAATTCCAAAGGGAGGAGGGTATAATGAAGCATGTCCAATTCCCCATGCCCCACCCCCACCCCCACATCCTAT AATGGCCTGAACTAGTTTTTCAGGTTAACTTTGGAATACCCTTGGCCAAGAAGAGGGGTCCATGCAGATGTTTGGAGGGGGGGGTCTTAA AGCTTTATTTTTAGATTACAGCATATTGCTTTTGCACTATCATAAAGTAAAAACATCATAAGTCAAGCCACCATAAATAGCGGACCATCT GTATATCTGAAAGAAGATGAACCAAGACTTGGTTATTGTTAAAGCTGATGATTGGCTGTATGAAGATTTACTATACAATTTTGTGTACTT TTGTAAGTTCTTGGTTGTTTCTAAACTTATAACAAAATAAATAATTGGCCATCTTCATTGGATTGTGAAGCCATTTCCTCCCACATGTTC GTAAAAATGGTAGCCATGGCCCCTGACTGAACCCAGGTATGAATGGCCTTCATTGATTTTGTAGACCCTGACCTTCTCTGTTCTTCCAGA GAAGGTGGTACTCAGATAAAAACTTTGAAGCATTTTATTGTTCAGAATATGACTTTGATCTTCTCATAAGACTCAAGTTTTTGAATGCTT AAGGCTCTTTATCGCTAGTGTATGTAAGTAATATCATTGTCCTTAACCACCATCTGCCTCTCCCCCCAGACACACATAAGTTTATGATAA CTACAGAGGTGTCCAGATGAGTAAAAACTGCCTTTGCATTAAGGTGAGAGAAGACCGAGATGAAATCTTATTAGAAAATATAACTGGGGA AAGCAGTTTGTCTGTTTCTCCTGGAAAACTACTAGACTACATCAACTTATATACTACTTCTTTTCTATATTCTCAATATGAGCCACTTAA GAAAATTAATGAGACTTTATGTTTGACCAAAGCTGGTAATTCCTTGATCTTATTCTCCAAACTTGATTCTGAATGACTACTGACCACTAA AAACACTAAGAGTACCACTGTGGATGTTTAAAAACATACACCCTAAGAGCCTTCAGAATTAGACGCTTCAATTCTTCTGAAATCAAAAGT CACACCAGCTTTGCTGTAGTTAATCAGGTAATACCTTTCTTCTCCACTACTCTCTAATTTCCTTGAGGGCAAATATTGTCTTTCTTGATA AATGCCCTGGACCCAACATCTAACAGAATACCTGGCATAAAGTGAAATCCATCGAATGTGCTAATGATAAATAAAGAAGTTCAAAAAAAT CTTTTAATAGAAGCTATAAAATAGCAGATAAGCTAAGTCATTCTCATAAAACACCATTTGTCATTTGAATGCGTGCATTGTGGCCTGTTA CTTTTAACTAGTCTCACTAATTTATAGTTATATATGATGTAGATCTAGATTGTGATGTACACTAAGTGGGTTGATCCTGAGATCAAGCTA TGATTGCTGCTTGCGTAAAGTGTTCCTTTTGGGAAATAAATAATCTTTCATATCTGTAAACTTTGGTATAATTGGTTATTTATGCAATGT ATTGTTGTGGTTGTCAACTCAAGATTGTATTCTCATCTGGGGACATTATGAATCTTGTTATTTTATCCCCTTTTTTGCTCTGTTTTTTCA CATTTATGGCCTACCTGTTACAGCTGCACGGTATCCATTTTCTGTGCTAAATTCATCTTTAATTTATTATTTTTTTTCAACCTCAGCTTT TAACTTTGCCCTGATTTTCTCTTCCATTTTATTTTCACTTCCATTTTAAGCTGGCTCAAAGCCTTTTAGGAACAATAAGTTACTGAATTA TATGAACCTATTTGAGGTAATGATGATGCCTGGGAAGGAGCAGTATTGCTGGATCAGAATTCACTTTTTCCAAGTTACAGATTTTAATAA ACCACTGTGCAAAGCATTAGGATAAATTTTTATTGAAAATTGAGTGAATGACTGAACAAATCGCTGGGTAATGCTGTTTAGGTTGTTCTA TTTTCCACGGATTAAATTAAATTCTATGTA >25589_25589_1_EHMT1-SPATA17_EHMT1_chr9_140513501_ENST00000460843_SPATA17_chr1_217856600_ENST00000366933_length(amino acids)=276AA_BP=12 MRARAMAAADAEIQRRWRGYRVRKYLFNYYYLKEYLKVVSETNDAIRKALEEFAEMKEREEKKANLEREEKKRDYQARKMHYLLSTKQIP GIYNSPFRKEPDPWELQLQKAKPLTHRRPKVKQKDSTSLTDWLACTSARSFPRSEILPPINRKQCQGPFRDITEVLEQRYRPLEPTLRVA EPIDELKLAREELRREEWLQNVNDNMFLPFSSYHKNEKYIPSMHLSSKYGPISYKEQFRSENPKKWICDKDFQTVLPSFELFSKYGKLYS KAGQIV -------------------------------------------------------------- >25589_25589_2_EHMT1-SPATA17_EHMT1_chr9_140513501_ENST00000462484_SPATA17_chr1_217856600_ENST00000366933_length(transcript)=5530nt_BP=58nt GCGCGGGAGGGGCGGGGCCACGCTGCGGGCCCGGGCCATGGCCGCCGCCGATGCCGAGATTCAGAGACGATGGCGAGGCTATAGGGTTCG GAAGTACCTCTTTAATTATTATTATTTGAAAGAGTACCTGAAAGTCGTTTCAGAGACCAATGATGCAATTAGGAAGGCACTGGAGGAGTT TGCAGAAATGAAAGAAAGAGAAGAGAAGAAGGCTAACCTCGAAAGGGAAGAGAAGAAAAGAGATTACCAAGCCCGAAAGATGCATTACCT CCTCAGCACAAAGCAGATTCCAGGAATATACAATTCACCCTTCAGAAAAGAGCCTGATCCATGGGAGCTGCAATTACAGAAGGCAAAGCC TTTAACACACCGAAGACCTAAAGTTAAGCAGAAGGACTCCACCAGCCTTACTGATTGGCTAGCTTGTACAAGCGCCCGTTCTTTTCCTCG GTCTGAAATTCTACCACCTATTAATAGAAAGCAATGTCAGGGGCCCTTCCGAGATATCACCGAAGTATTAGAACAACGCTACAGGCCTTT GGAGCCAACGTTGCGGGTGGCAGAACCAATCGATGAGTTAAAGTTGGCCAGAGAGGAGCTCAGAAGAGAGGAATGGCTGCAAAATGTAAA TGACAATATGTTTTTGCCATTTTCTTCATACCATAAAAATGAAAAGTACATCCCATCAATGCATTTATCAAGCAAGTATGGTCCTATTTC TTACAAAGAACAATTCCGAAGTGAAAATCCTAAGAAATGGATCTGTGACAAGGATTTCCAGACTGTATTACCATCATTTGAGCTCTTCTC AAAGTATGGAAAATTATATTCAAAAGCTGGACAGATTGTATAAAGGCGTCAGAAGAAGAAACTGAAGCCATCTGCATTTTAAAACTTAAC AGTTCTGAAAGGAAAACACAGATGAAGATCCTGTAGGAAATATACTTGCTATGATTCAATAAACTATAAAATTTTGTAGCTCTAGTCATG AATTAATTATTGTGTGTATGTATTTGAAATCTCTTAGACATATCTTTTCAAACAAATACTTAAACTTGAGTCTGTTACCTCTGAGAATGT CTGTCACAATCACAGCCCCTGGTGACCAACAGTGCCATAAATGGTTCCCCTGAAATACCCATATTCTGACCCCATCACCCGGGTCACAAA TGATGACACCAGTGCTGGGTACCCAAATTAAGGGCGACTAACATGTAAGTAGGATGATGAATGGTTTGAAGTGTAAATTGTGTGAAATAG TTCAGCATGAAAGCTCTGCCCTGAGAAATAATGACTGATTATACCAGTTAGAAACCCTTCAGACTGTTGAACTTGAGACACACAGACACA ACAGACACAGACTAGTGGGTGTGGGAGCAGAAGCATAGGCACTCTCAAATAGAGAAGACATTTGCAAGTCTCTGCTGCCAAGGGAGCAAA GAGATAATCTCGATGCCAGGGCTGTGTCAGATCCTGCATGGCTCTGAACTACAGTTGGTTTCCAAAAAGCTGGGCTTTTGTGAGGTTTCT GTTTTTTATGCGATGCTTAGATTTTCCTGGAATCCTTGGAAGTTAACTCAACATTTGAGGTTCCTATATCCTTTAATCCCATCCCATGTG TATCTTTACAATAAAACTTCCTGCTAGAGGTATCCTGAGGGAATCTTTGTTTCTTGCATCTGAAGAGAATAACACTAAACTTATTGCATA TGATTTGGGGGTTGGATCCAATGATAGCACAGAGGGACAGGAAATGTGATTTATTGACCATAAAAGCTTTAATTTAAAATTGCTGGCCAG CGAAATCCAGACTCTGGGAAATGTTATAGGCCAAATGATCTGTTTCTTCAACAAATAAATTACAAAACTGAGGGGAAAAAAAGGAACCTA TTAAAAAGACATTATCTATCAATCACTGTGTGTGTACAGTTTTGAGCTCCTGATTTAAATAAACAAATTGTAAAATAAAAGATCTTTTGA AACAATTGGAAGTTTGAACACTGATTGGATATTGGTGGTATAAAAGAATTATTGGTTTTTTTTAGGTGTCATGGTATTGTTTTGGTTTAC AAAGTTATAGATATAAATGTTGAAATATTTACAGATGCAGTGATATGTTGGCTGGGATTTAATTTGAAAGTCAAAGAAGTGAGTTAAAAT ACATATACTCCTCCTCTTATAATCGGTTATGTGCCAATAAATCCATTGTAAATTGAAAATATCCTAAGTTGAAAATATTGTAAGTTAAAA ATGCATTTAATACAGCTAACTTACCAGACAGCTTAGCTTAGCCTCACCTACCTTAAATATGCTCAGAACACATACGTTAGCCTATAGTTG GGCAAAATCATCTAACACAAAGCCTATTTTATAATATTGAACAGCCCATGTAATTTATTGAATATTGTACTAAAAGTGAAAAACTGAATA GTTGTATGCATACTCAAAGTATGGTTTCTACTGAATGCATTGTAAACCAGAAAGTATCTGAGACAATGTTTTTTTTTTTTTTTTTTTTTT TTTTAATTTCTGAGACAGAGTCTCATTCTGTCACCCAGGTTGGAGTGCAGTGGCATGAACTCAGCTCACTGCAACCTTCACCTCCCAGGC TCAAGCAGTCCTCCCATCTCAGCCTCCCAAGTAGCTGGGACCACAGGCATGGGCCAACAAGCCTGGCTAAATATTTTGTACTTTTGGTAG ACACGGGTTTTTACCATGTTGTGCAGGCTGGTCTTGAACGCCTGAGCTCAAGTGATCTGCCTGCCTCGGCCTCCCAAAGTACTGGGATTA CAGGCATGAGTCACTGAACCCAGCCCTGAGACATGTCTCAATTAGTTTAGAAGTTTATTTGGCCAAGGTTAAGGACATGCCCTTGACACT GCACCTGTCAGTACAGGTCCTGACAACATGTGTCCAAGGTGGTCAAGCTATACAACTTGGTTTTATACATTTTAAGGAGACATAAGACAT CAATAAATACATGGAAGATGTACATTGGTTCAGTCTGGAAAGGCAGGATGCCTCAAACTTGAAACAAGAGAATGGGGGGTGGGAGGATGT GGGAGATGCTTCCAGGTCATAGTTGGAGTCAAAGACTTTCCAATTGGCAATTGGTTCAAAGAATTTATCTAAAGACCTGGAATCCATAGA AGGCTTTACCTGGATTAAAATAAGGGGACCAAGGTTCTCATGCAGATGAAGCCTCCAGGTGGCAGACTTCAGAGAGCATAGATTGTAAAT GTTTCTTATCTGACTTAAAAAGGTGTCAGACTCTTAGTAAATTATCTCCTGGATCAGTGAAAAGATGTGAAAAAAGGTTCTCTACAGAAT GTAGATTTTCCCCATAAGAGATAGCTTCGCAGGGCCATTGCAAAATATGTCAAAAATATATATTTTAGGGTAAAATGCCTCAATTTATTT CAAGGCCTGATATTTGTCATGTGATGCTATACTACAGTCAGGCTGAAACTTGGCATCTTATTGCTATGAAAAATCTTAAGATCTCTATTT TAATGTTAATGCTGGTCACTGGTGCCTGAATTCCAAAGGGAGGAGGGTATAATGAAGCATGTCCAATTCCCCATGCCCCACCCCCACCCC CACATCCTATAATGGCCTGAACTAGTTTTTCAGGTTAACTTTGGAATACCCTTGGCCAAGAAGAGGGGTCCATGCAGATGTTTGGAGGGG GGGGTCTTAAAGCTTTATTTTTAGATTACAGCATATTGCTTTTGCACTATCATAAAGTAAAAACATCATAAGTCAAGCCACCATAAATAG CGGACCATCTGTATATCTGAAAGAAGATGAACCAAGACTTGGTTATTGTTAAAGCTGATGATTGGCTGTATGAAGATTTACTATACAATT TTGTGTACTTTTGTAAGTTCTTGGTTGTTTCTAAACTTATAACAAAATAAATAATTGGCCATCTTCATTGGATTGTGAAGCCATTTCCTC CCACATGTTCGTAAAAATGGTAGCCATGGCCCCTGACTGAACCCAGGTATGAATGGCCTTCATTGATTTTGTAGACCCTGACCTTCTCTG TTCTTCCAGAGAAGGTGGTACTCAGATAAAAACTTTGAAGCATTTTATTGTTCAGAATATGACTTTGATCTTCTCATAAGACTCAAGTTT TTGAATGCTTAAGGCTCTTTATCGCTAGTGTATGTAAGTAATATCATTGTCCTTAACCACCATCTGCCTCTCCCCCCAGACACACATAAG TTTATGATAACTACAGAGGTGTCCAGATGAGTAAAAACTGCCTTTGCATTAAGGTGAGAGAAGACCGAGATGAAATCTTATTAGAAAATA TAACTGGGGAAAGCAGTTTGTCTGTTTCTCCTGGAAAACTACTAGACTACATCAACTTATATACTACTTCTTTTCTATATTCTCAATATG AGCCACTTAAGAAAATTAATGAGACTTTATGTTTGACCAAAGCTGGTAATTCCTTGATCTTATTCTCCAAACTTGATTCTGAATGACTAC TGACCACTAAAAACACTAAGAGTACCACTGTGGATGTTTAAAAACATACACCCTAAGAGCCTTCAGAATTAGACGCTTCAATTCTTCTGA AATCAAAAGTCACACCAGCTTTGCTGTAGTTAATCAGGTAATACCTTTCTTCTCCACTACTCTCTAATTTCCTTGAGGGCAAATATTGTC TTTCTTGATAAATGCCCTGGACCCAACATCTAACAGAATACCTGGCATAAAGTGAAATCCATCGAATGTGCTAATGATAAATAAAGAAGT TCAAAAAAATCTTTTAATAGAAGCTATAAAATAGCAGATAAGCTAAGTCATTCTCATAAAACACCATTTGTCATTTGAATGCGTGCATTG TGGCCTGTTACTTTTAACTAGTCTCACTAATTTATAGTTATATATGATGTAGATCTAGATTGTGATGTACACTAAGTGGGTTGATCCTGA GATCAAGCTATGATTGCTGCTTGCGTAAAGTGTTCCTTTTGGGAAATAAATAATCTTTCATATCTGTAAACTTTGGTATAATTGGTTATT TATGCAATGTATTGTTGTGGTTGTCAACTCAAGATTGTATTCTCATCTGGGGACATTATGAATCTTGTTATTTTATCCCCTTTTTTGCTC TGTTTTTTCACATTTATGGCCTACCTGTTACAGCTGCACGGTATCCATTTTCTGTGCTAAATTCATCTTTAATTTATTATTTTTTTTCAA CCTCAGCTTTTAACTTTGCCCTGATTTTCTCTTCCATTTTATTTTCACTTCCATTTTAAGCTGGCTCAAAGCCTTTTAGGAACAATAAGT TACTGAATTATATGAACCTATTTGAGGTAATGATGATGCCTGGGAAGGAGCAGTATTGCTGGATCAGAATTCACTTTTTCCAAGTTACAG ATTTTAATAAACCACTGTGCAAAGCATTAGGATAAATTTTTATTGAAAATTGAGTGAATGACTGAACAAATCGCTGGGTAATGCTGTTTA GGTTGTTCTATTTTCCACGGATTAAATTAAATTCTATGTA >25589_25589_2_EHMT1-SPATA17_EHMT1_chr9_140513501_ENST00000462484_SPATA17_chr1_217856600_ENST00000366933_length(amino acids)=276AA_BP=12 MRARAMAAADAEIQRRWRGYRVRKYLFNYYYLKEYLKVVSETNDAIRKALEEFAEMKEREEKKANLEREEKKRDYQARKMHYLLSTKQIP GIYNSPFRKEPDPWELQLQKAKPLTHRRPKVKQKDSTSLTDWLACTSARSFPRSEILPPINRKQCQGPFRDITEVLEQRYRPLEPTLRVA EPIDELKLAREELRREEWLQNVNDNMFLPFSSYHKNEKYIPSMHLSSKYGPISYKEQFRSENPKKWICDKDFQTVLPSFELFSKYGKLYS KAGQIV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for EHMT1-SPATA17 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1162_1181 | 0 | 826.0 | histone H3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000334856 | + | 1 | 17 | 1242_1245 | 0 | 826.0 | histone H3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1162_1181 | 7.0 | 1299.0 | histone H3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000460843 | + | 1 | 27 | 1242_1245 | 7.0 | 1299.0 | histone H3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1162_1181 | 7.0 | 809.0 | histone H3 |
| Hgene | EHMT1 | chr9:140513501 | chr1:217856600 | ENST00000462484 | + | 1 | 16 | 1242_1245 | 7.0 | 809.0 | histone H3 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for EHMT1-SPATA17 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for EHMT1-SPATA17 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | EHMT1 | C0795833 | KLEEFSTRA SYNDROME 1 | 11 | CLINGEN;CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | EHMT1 | C0025149 | Medulloblastoma | 1 | CTD_human |
| Hgene | EHMT1 | C0205833 | Medullomyoblastoma | 1 | CTD_human |
| Hgene | EHMT1 | C0278510 | Childhood Medulloblastoma | 1 | CTD_human |
| Hgene | EHMT1 | C0278876 | Adult Medulloblastoma | 1 | CTD_human |
| Hgene | EHMT1 | C0751291 | Desmoplastic Medulloblastoma | 1 | CTD_human |
| Hgene | EHMT1 | C1275668 | Melanotic medulloblastoma | 1 | CTD_human |
