
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FCRL5-PHF11 (FusionGDB2 ID:HG83416TG51131) |
Fusion Gene Summary for FCRL5-PHF11 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FCRL5-PHF11 | Fusion gene ID: hg83416tg51131 | Hgene | Tgene | Gene symbol | FCRL5 | PHF11 | Gene ID | 83416 | 51131 |
| Gene name | Fc receptor like 5 | PHD finger protein 11 | |
| Synonyms | BXMAS1|CD307|CD307e|FCRH5|IRTA2|PRO820 | APY|BCAP|IGEL|IGER|IGHER|NY-REN-34|NYREN34 | |
| Cytomap | ('FCRL5')('PHF11') 1q23.1 | 13q14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | Fc receptor-like protein 5fc receptor homolog 5fcR-like protein 5immune receptor translocation-associated protein 2immunoglobulin superfamily receptor translocation associated 2 (IRTA2) | PHD finger protein 11BRCA1 C-terminus-associated proteinIgE responsiveness (atopic)renal carcinoma antigen NY-REN-34 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000356953, ENST00000361835, ENST00000368190, ENST00000368191, ENST00000368188, ENST00000368189, ENST00000461387, | ||
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 4 X 5 X 3=60 |
| # samples | 2 | 8 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(8/60*10)=0.415037499278844 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: FCRL5 [Title/Abstract] AND PHF11 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FCRL5(157504404)-PHF11(50080771), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FCRL5-PHF11 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FCRL5-PHF11 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FCRL5-PHF11 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. FCRL5-PHF11 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across FCRL5 (5'-gene) Fusion gene breakpoints across FCRL5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
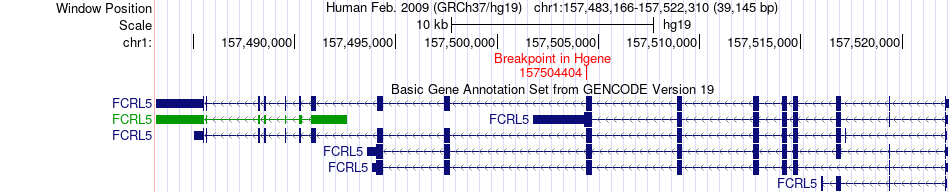 |
 Fusion gene breakpoints across PHF11 (3'-gene) Fusion gene breakpoints across PHF11 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
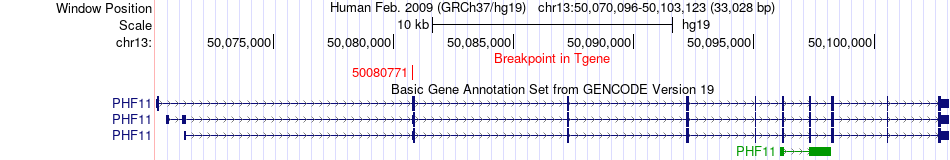 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GT-01A | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
Top |
Fusion Gene ORF analysis for FCRL5-PHF11 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000356953 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000356953 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000361835 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000361835 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000368190 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000368190 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000368191 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-5UTR | ENST00000368191 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-intron | ENST00000356953 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-intron | ENST00000361835 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-intron | ENST00000368190 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| 5CDS-intron | ENST00000368191 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| In-frame | ENST00000356953 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| In-frame | ENST00000361835 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| In-frame | ENST00000368190 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| In-frame | ENST00000368191 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-3CDS | ENST00000368188 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-3CDS | ENST00000368189 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-3CDS | ENST00000461387 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000368188 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000368188 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000368189 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000368189 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000461387 | ENST00000357596 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-5UTR | ENST00000461387 | ENST00000488958 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-intron | ENST00000368188 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-intron | ENST00000368189 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
| intron-intron | ENST00000461387 | ENST00000460489 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361835 | FCRL5 | chr1 | 157504404 | - | ENST00000378319 | PHF11 | chr13 | 50080771 | + | 3063 | 1839 | 140 | 2740 | 866 |
| ENST00000356953 | FCRL5 | chr1 | 157504404 | - | ENST00000378319 | PHF11 | chr13 | 50080771 | + | 2933 | 1709 | 10 | 2610 | 866 |
| ENST00000368190 | FCRL5 | chr1 | 157504404 | - | ENST00000378319 | PHF11 | chr13 | 50080771 | + | 2998 | 1774 | 75 | 2675 | 866 |
| ENST00000368191 | FCRL5 | chr1 | 157504404 | - | ENST00000378319 | PHF11 | chr13 | 50080771 | + | 2781 | 1557 | 113 | 2458 | 781 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361835 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + | 0.000167075 | 0.99983287 |
| ENST00000356953 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + | 0.000200576 | 0.99979943 |
| ENST00000368190 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + | 0.000162089 | 0.9998379 |
| ENST00000368191 | ENST00000378319 | FCRL5 | chr1 | 157504404 | - | PHF11 | chr13 | 50080771 | + | 0.000468455 | 0.9995315 |
Top |
Fusion Genomic Features for FCRL5-PHF11 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FCRL5 | chr1 | 157504403 | - | PHF11 | chr13 | 50080770 | + | 6.72E-05 | 0.9999329 |
| FCRL5 | chr1 | 157504403 | - | PHF11 | chr13 | 50080770 | + | 6.72E-05 | 0.9999329 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
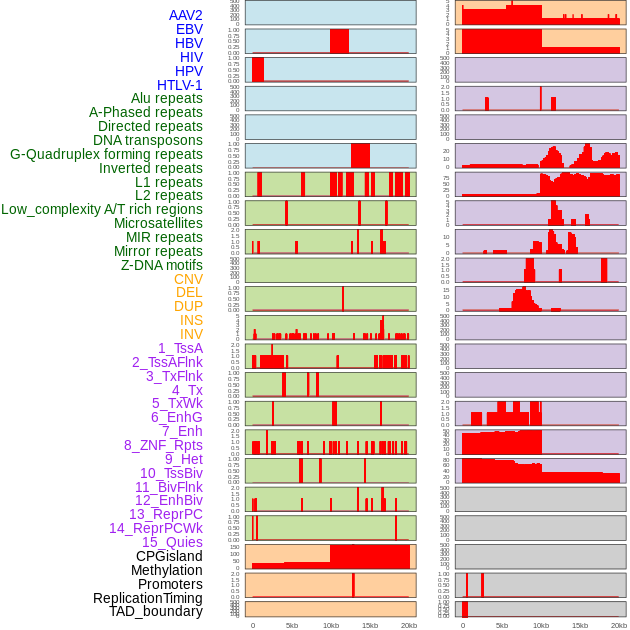 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
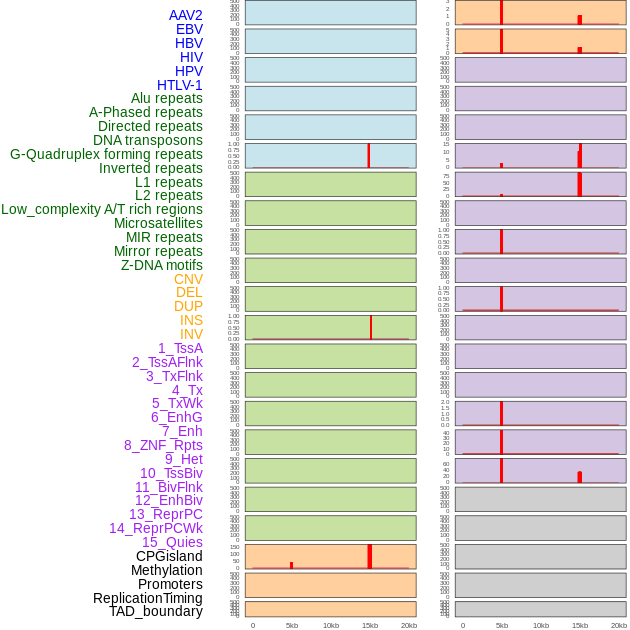 |
Top |
Fusion Protein Features for FCRL5-PHF11 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:157504404/chr13:50080771) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 188_271 | 560 | 978.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 23_101 | 560 | 978.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 287_374 | 560 | 978.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 380_463 | 560 | 978.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 473_556 | 560 | 978.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 188_271 | 560 | 760.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 23_101 | 560 | 760.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 287_374 | 560 | 760.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 380_463 | 560 | 760.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 473_556 | 560 | 760.0 | Domain | Note=Ig-like C2-type 5 |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000357596 | 1 | 11 | 108_160 | 0 | 293.0 | Zinc finger | PHD-type | |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000357596 | 1 | 11 | 42_78 | 0 | 293.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000378319 | 0 | 10 | 108_160 | 31 | 332.0 | Zinc finger | PHD-type | |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000378319 | 0 | 10 | 42_78 | 31 | 332.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000488958 | 0 | 10 | 108_160 | 0 | 293.0 | Zinc finger | PHD-type | |
| Tgene | PHF11 | chr1:157504404 | chr13:50080771 | ENST00000488958 | 0 | 10 | 42_78 | 0 | 293.0 | Zinc finger | C2HC pre-PHD-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 566_651 | 560 | 978.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 659_744 | 560 | 978.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 752_834 | 560 | 978.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 188_271 | 0 | 125.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 23_101 | 0 | 125.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 287_374 | 0 | 125.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 380_463 | 0 | 125.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 473_556 | 0 | 125.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 566_651 | 0 | 125.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 659_744 | 0 | 125.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 752_834 | 0 | 125.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 188_271 | 0 | 593.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 23_101 | 0 | 593.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 287_374 | 0 | 593.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 380_463 | 0 | 593.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 473_556 | 0 | 593.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 566_651 | 0 | 593.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 659_744 | 0 | 593.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 752_834 | 0 | 593.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 566_651 | 560 | 760.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 659_744 | 560 | 760.0 | Domain | Note=Ig-like C2-type 7 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 752_834 | 560 | 760.0 | Domain | Note=Ig-like C2-type 8 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 897_902 | 560 | 978.0 | Motif | Note=ITIM motif 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 910_915 | 560 | 978.0 | Motif | Note=ITIM motif 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 922_927 | 560 | 978.0 | Motif | Note=ITIM motif 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 952_957 | 560 | 978.0 | Motif | Note=ITIM motif 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 897_902 | 0 | 125.0 | Motif | Note=ITIM motif 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 910_915 | 0 | 125.0 | Motif | Note=ITIM motif 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 922_927 | 0 | 125.0 | Motif | Note=ITIM motif 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 952_957 | 0 | 125.0 | Motif | Note=ITIM motif 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 897_902 | 0 | 593.0 | Motif | Note=ITIM motif 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 910_915 | 0 | 593.0 | Motif | Note=ITIM motif 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 922_927 | 0 | 593.0 | Motif | Note=ITIM motif 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 952_957 | 0 | 593.0 | Motif | Note=ITIM motif 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 897_902 | 560 | 760.0 | Motif | Note=ITIM motif 1 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 910_915 | 560 | 760.0 | Motif | Note=ITIM motif 2 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 922_927 | 560 | 760.0 | Motif | Note=ITIM motif 3 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 952_957 | 560 | 760.0 | Motif | Note=ITIM motif 4 |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 16_851 | 560 | 978.0 | Topological domain | Extracellular |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 873_977 | 560 | 978.0 | Topological domain | Cytoplasmic |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 16_851 | 0 | 125.0 | Topological domain | Extracellular |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 873_977 | 0 | 125.0 | Topological domain | Cytoplasmic |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 16_851 | 0 | 593.0 | Topological domain | Extracellular |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 873_977 | 0 | 593.0 | Topological domain | Cytoplasmic |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 16_851 | 560 | 760.0 | Topological domain | Extracellular |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 873_977 | 560 | 760.0 | Topological domain | Cytoplasmic |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000361835 | - | 8 | 17 | 852_872 | 560 | 978.0 | Transmembrane | Helical |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368188 | - | 1 | 4 | 852_872 | 0 | 125.0 | Transmembrane | Helical |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368189 | - | 1 | 8 | 852_872 | 0 | 593.0 | Transmembrane | Helical |
| Hgene | FCRL5 | chr1:157504404 | chr13:50080771 | ENST00000368190 | - | 8 | 10 | 852_872 | 560 | 760.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FCRL5-PHF11 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >30034_30034_1_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000356953_PHF11_chr13_50080771_ENST00000378319_length(transcript)=2933nt_BP=1709nt ATCTCAGCCCTTGGTGGTCCAGGTCTTCATGCTGCTGTGGGTGATATTACTGGTCCTGGTGCCATCCCCTGGGCAATTTCCAAGGACACC CAGGCCCATTATTTTCCTCCAGCCTCCATGGACCACAGTCTTCCAAGGAGAGAGAGTGACCCTCACTTGCAAGGGATTTCGCTTCTACTC ACCACAGAAAACAAAATGGTACCATCGGTACCTTGGGAAAGAAATACTAAGAGAAACCCCAGACAATATCCTTGAGGTTCAGGAATCTGG AGAGTACAGATGCCAGGCCCAGGGCTCCCCTCTCAGTAGCCCTGTGCACTTGGATTTTTCTTCAGCTTCGCTGATCCTGCAAGCTCCACT TTCTGTGTTTGAAGGAGACTCTGTGGTTCTGAGGTGCCGGGCAAAGGCGGAAGTAACACTGAATAATACTATTTACAAGAATGATAATGT CCTGGCATTCCTTAATAAAAGAACTGACTTCCATATTCCTCATGCATGTCTCAAGGACAATGGTGCATATCGCTGTACTGGATATAAGGA AAGTTGTTGCCCTGTTTCTTCCAATACAGTCAAAATCCAAGTCCAAGAGCCATTTACACGTCCAGTGCTGAGAGCCAGCTCCTTCCAGCC CATCAGCGGGAACCCAGTGACCCTGACCTGTGAGACCCAGCTCTCTCTAGAGAGGTCAGATGTCCCGCTCCGGTTCCGCTTCTTCAGAGA TGACCAGACCCTGGGATTAGGCTGGAGTCTCTCCCCGAATTTCCAGATTACTGCCATGTGGAGTAAAGATTCAGGGTTCTACTGGTGTAA GGCAGCAACAATGCCTTACAGCGTCATATCTGACAGCCCGAGATCCTGGATACAGGTGCAGATCCCTGCATCTCATCCTGTCCTCACTCT CAGCCCTGAAAAGGCTCTGAATTTTGAGGGAACCAAGGTGACACTTCACTGTGAAACCCAGGAAGATTCTCTGCGCACTTTGTACAGGTT TTATCATGAGGGTGTCCCCCTGAGGCACAAGTCAGTCCGCTGTGAAAGGGGAGCATCCATCAGCTTCTCACTGACTACAGAGAATTCAGG GAACTACTACTGCACAGCTGACAATGGCCTTGGCGCCAAGCCCAGTAAGGCTGTGAGCCTCTCAGTCACTGTTCCCGTGTCTCATCCTGT CCTCAACCTCAGCTCTCCTGAGGACCTGATTTTTGAGGGAGCCAAGGTGACACTTCACTGTGAAGCCCAGAGAGGTTCACTCCCCATCCT GTACCAGTTTCATCATGAGGGTGCTGCCCTGGAGCGTAGGTCGGCCAACTCTGCAGGAGGAGTGGCCATCAGCTTCTCTCTGACTGCAGA GCATTCAGGGAACTACTACTGCACAGCTGACAATGGCTTTGGCCCCCAGCGCAGTAAGGCGGTGAGCCTCTCCGTCACTGTCCCTGTGTC TCATCCTGTCCTCACCCTCAGCTCTGCTGAGGCCCTGACTTTTGAAGGAGCCACTGTGACACTTCACTGTGAAGTCCAGAGAGGTTCCCC ACAAATCCTATACCAGTTTTATCATGAGGACATGCCCCTGTGGAGCAGCTCAACACCCTCTGTGGGAAGAGTGTCCTTCAGCTTCTCTCT GACTGAAGGACATTCAGGGAATTACTACTGCACAGCTGACAATGGCTTTGGTCCCCAGCGCAGTGAAGTGGTGAGCCTTTTTGTCACTGG TGTCTTTCAGGTTGCAGAAAAGATGGAAAAAAGGACATGTGCACTCTGCCCCAAAGATGTCGAATATAATGTCCTATACTTTGCACAATC AGAGAATATAGCTGCTCATGAGAATTGTTTGCTGTATTCTTCAGGACTTGTGGAATGTGAGGATCAGGATCCACTTAATCCTGATAGAAG TTTTGATGTGGAATCAGTAAAGAAAGAAATCCAGAGAGGAAGGAAGTTGAAATGCAAATTTTGTCATAAAAGAGGAGCCACCGTGGGATG TGATTTAAAAAACTGTAACAAGAATTACCACTTTTTCTGTGCCAAGAAGGACGACGCAGTTCCACAGTCTGATGGAGTTCGAGGAATTTA TAAACTGCTTTGCCAGCAACATGCTCAATTCCCGATCATCGCTCAAAGTGCTAAATTTTCAGGAGTGAAAAGAAAAAGAGGAAGGAAGAA ACCCCTCTCAGGCAATCATGTACAGCCACCCGAAACAATGAAATGTAATACATTCATAAGACAAGTGAAAGAAGAGCATGGCAGACACAC AGATGCAACTGTGAAAGTTCCTTTTCTTAAGAAATGCAAGGAAGCAGGACTTCTTAATTACTTACTTGAAGAAATATTAGACAAAGTTCA TTCAATTCCAGAAAAACTCATGGATGAGACTACTTCAGAATCAGACTATGAAGAAATCGGGAGTGCACTTTTTGACTGTAGATTGTTCGA AGACACATTTGTAAATTTTCAAGCAGCAATAGAGAAAAAAATTCATGCATCTCAACAAAGGTGGCAGCAGTTGAAGGAAGAGATTGAGCT ACTTCAGGACTTAAAACAAACCTTGTGCTCTTTTCAAGAAAATAGAGATCTTATGTCAAGTTCTACATCAATATCATCCCTGTCTTATTA GGGATTACCGTTTCCTAAGCCAAGAGTCATGTCAAATTGCAATCAGGCTCAAAACCAGAGACCAGGCTGTGAAATCCACACATCTTTAGA ACTAGTCGTCTCCTCTTGGCCTCAGCAGCTCTTCCCTGTTCTTACTGGTTGACATTTTGATCACTCTTTGCACACTCTTGTGTTTTTTGC TCACTGTCACATTCCCAGCACCTAGTATGCTCAGTAAATGTTTGTGGAATAAGTGCATAAAATGTTCTTAACCTTTGATTCTACTTACAG CCCATGATAGCCTCTTAGATATAATAAATTTGGATTATACTACTTTACTTGTA >30034_30034_1_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000356953_PHF11_chr13_50080771_ENST00000378319_length(amino acids)=866AA_BP=566 MVVQVFMLLWVILLVLVPSPGQFPRTPRPIIFLQPPWTTVFQGERVTLTCKGFRFYSPQKTKWYHRYLGKEILRETPDNILEVQESGEYR CQAQGSPLSSPVHLDFSSASLILQAPLSVFEGDSVVLRCRAKAEVTLNNTIYKNDNVLAFLNKRTDFHIPHACLKDNGAYRCTGYKESCC PVSSNTVKIQVQEPFTRPVLRASSFQPISGNPVTLTCETQLSLERSDVPLRFRFFRDDQTLGLGWSLSPNFQITAMWSKDSGFYWCKAAT MPYSVISDSPRSWIQVQIPASHPVLTLSPEKALNFEGTKVTLHCETQEDSLRTLYRFYHEGVPLRHKSVRCERGASISFSLTTENSGNYY CTADNGLGAKPSKAVSLSVTVPVSHPVLNLSSPEDLIFEGAKVTLHCEAQRGSLPILYQFHHEGAALERRSANSAGGVAISFSLTAEHSG NYYCTADNGFGPQRSKAVSLSVTVPVSHPVLTLSSAEALTFEGATVTLHCEVQRGSPQILYQFYHEDMPLWSSSTPSVGRVSFSFSLTEG HSGNYYCTADNGFGPQRSEVVSLFVTGVFQVAEKMEKRTCALCPKDVEYNVLYFAQSENIAAHENCLLYSSGLVECEDQDPLNPDRSFDV ESVKKEIQRGRKLKCKFCHKRGATVGCDLKNCNKNYHFFCAKKDDAVPQSDGVRGIYKLLCQQHAQFPIIAQSAKFSGVKRKRGRKKPLS GNHVQPPETMKCNTFIRQVKEEHGRHTDATVKVPFLKKCKEAGLLNYLLEEILDKVHSIPEKLMDETTSESDYEEIGSALFDCRLFEDTF VNFQAAIEKKIHASQQRWQQLKEEIELLQDLKQTLCSFQENRDLMSSSTSISSLSY -------------------------------------------------------------- >30034_30034_2_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000361835_PHF11_chr13_50080771_ENST00000378319_length(transcript)=3063nt_BP=1839nt AATTCACTAATGCATTCTGCTCTTTTTGAGAGCACAGCTTCTCAGATGTGCTCCTTGGAGCTGGTGTGCAGTGTCCTGACTGTAAGATCA AGTCCAAACCTGTTTTGGAATTGAGGAAACTTCTCTTTTGATCTCAGCCCTTGGTGGTCCAGGTCTTCATGCTGCTGTGGGTGATATTAC TGGTCCTGGCTCCTGTCAGTGGACAGTTTGCAAGGACACCCAGGCCCATTATTTTCCTCCAGCCTCCATGGACCACAGTCTTCCAAGGAG AGAGAGTGACCCTCACTTGCAAGGGATTTCGCTTCTACTCACCACAGAAAACAAAATGGTACCATCGGTACCTTGGGAAAGAAATACTAA GAGAAACCCCAGACAATATCCTTGAGGTTCAGGAATCTGGAGAGTACAGATGCCAGGCCCAGGGCTCCCCTCTCAGTAGCCCTGTGCACT TGGATTTTTCTTCAGCTTCGCTGATCCTGCAAGCTCCACTTTCTGTGTTTGAAGGAGACTCTGTGGTTCTGAGGTGCCGGGCAAAGGCGG AAGTAACACTGAATAATACTATTTACAAGAATGATAATGTCCTGGCATTCCTTAATAAAAGAACTGACTTCCATATTCCTCATGCATGTC TCAAGGACAATGGTGCATATCGCTGTACTGGATATAAGGAAAGTTGTTGCCCTGTTTCTTCCAATACAGTCAAAATCCAAGTCCAAGAGC CATTTACACGTCCAGTGCTGAGAGCCAGCTCCTTCCAGCCCATCAGCGGGAACCCAGTGACCCTGACCTGTGAGACCCAGCTCTCTCTAG AGAGGTCAGATGTCCCGCTCCGGTTCCGCTTCTTCAGAGATGACCAGACCCTGGGATTAGGCTGGAGTCTCTCCCCGAATTTCCAGATTA CTGCCATGTGGAGTAAAGATTCAGGGTTCTACTGGTGTAAGGCAGCAACAATGCCTTACAGCGTCATATCTGACAGCCCGAGATCCTGGA TACAGGTGCAGATCCCTGCATCTCATCCTGTCCTCACTCTCAGCCCTGAAAAGGCTCTGAATTTTGAGGGAACCAAGGTGACACTTCACT GTGAAACCCAGGAAGATTCTCTGCGCACTTTGTACAGGTTTTATCATGAGGGTGTCCCCCTGAGGCACAAGTCAGTCCGCTGTGAAAGGG GAGCATCCATCAGCTTCTCACTGACTACAGAGAATTCAGGGAACTACTACTGCACAGCTGACAATGGCCTTGGCGCCAAGCCCAGTAAGG CTGTGAGCCTCTCAGTCACTGTTCCCGTGTCTCATCCTGTCCTCAACCTCAGCTCTCCTGAGGACCTGATTTTTGAGGGAGCCAAGGTGA CACTTCACTGTGAAGCCCAGAGAGGTTCACTCCCCATCCTGTACCAGTTTCATCATGAGGGTGCTGCCCTGGAGCGTAGGTCGGCCAACT CTGCAGGAGGAGTGGCCATCAGCTTCTCTCTGACTGCAGAGCATTCAGGGAACTACTACTGCACAGCTGACAATGGCTTTGGCCCCCAGC GCAGTAAGGCGGTGAGCCTCTCCGTCACTGTCCCTGTGTCTCATCCTGTCCTCACCCTCAGCTCTGCTGAGGCCCTGACTTTTGAAGGAG CCACTGTGACACTTCACTGTGAAGTCCAGAGAGGTTCCCCACAAATCCTATACCAGTTTTATCATGAGGACATGCCCCTGTGGAGCAGCT CAACACCCTCTGTGGGAAGAGTGTCCTTCAGCTTCTCTCTGACTGAAGGACATTCAGGGAATTACTACTGCACAGCTGACAATGGCTTTG GTCCCCAGCGCAGTGAAGTGGTGAGCCTTTTTGTCACTGGTGTCTTTCAGGTTGCAGAAAAGATGGAAAAAAGGACATGTGCACTCTGCC CCAAAGATGTCGAATATAATGTCCTATACTTTGCACAATCAGAGAATATAGCTGCTCATGAGAATTGTTTGCTGTATTCTTCAGGACTTG TGGAATGTGAGGATCAGGATCCACTTAATCCTGATAGAAGTTTTGATGTGGAATCAGTAAAGAAAGAAATCCAGAGAGGAAGGAAGTTGA AATGCAAATTTTGTCATAAAAGAGGAGCCACCGTGGGATGTGATTTAAAAAACTGTAACAAGAATTACCACTTTTTCTGTGCCAAGAAGG ACGACGCAGTTCCACAGTCTGATGGAGTTCGAGGAATTTATAAACTGCTTTGCCAGCAACATGCTCAATTCCCGATCATCGCTCAAAGTG CTAAATTTTCAGGAGTGAAAAGAAAAAGAGGAAGGAAGAAACCCCTCTCAGGCAATCATGTACAGCCACCCGAAACAATGAAATGTAATA CATTCATAAGACAAGTGAAAGAAGAGCATGGCAGACACACAGATGCAACTGTGAAAGTTCCTTTTCTTAAGAAATGCAAGGAAGCAGGAC TTCTTAATTACTTACTTGAAGAAATATTAGACAAAGTTCATTCAATTCCAGAAAAACTCATGGATGAGACTACTTCAGAATCAGACTATG AAGAAATCGGGAGTGCACTTTTTGACTGTAGATTGTTCGAAGACACATTTGTAAATTTTCAAGCAGCAATAGAGAAAAAAATTCATGCAT CTCAACAAAGGTGGCAGCAGTTGAAGGAAGAGATTGAGCTACTTCAGGACTTAAAACAAACCTTGTGCTCTTTTCAAGAAAATAGAGATC TTATGTCAAGTTCTACATCAATATCATCCCTGTCTTATTAGGGATTACCGTTTCCTAAGCCAAGAGTCATGTCAAATTGCAATCAGGCTC AAAACCAGAGACCAGGCTGTGAAATCCACACATCTTTAGAACTAGTCGTCTCCTCTTGGCCTCAGCAGCTCTTCCCTGTTCTTACTGGTT GACATTTTGATCACTCTTTGCACACTCTTGTGTTTTTTGCTCACTGTCACATTCCCAGCACCTAGTATGCTCAGTAAATGTTTGTGGAAT AAGTGCATAAAATGTTCTTAACCTTTGATTCTACTTACAGCCCATGATAGCCTCTTAGATATAATAAATTTGGATTATACTACTTTACTT GTA >30034_30034_2_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000361835_PHF11_chr13_50080771_ENST00000378319_length(amino acids)=866AA_BP=566 MVVQVFMLLWVILLVLAPVSGQFARTPRPIIFLQPPWTTVFQGERVTLTCKGFRFYSPQKTKWYHRYLGKEILRETPDNILEVQESGEYR CQAQGSPLSSPVHLDFSSASLILQAPLSVFEGDSVVLRCRAKAEVTLNNTIYKNDNVLAFLNKRTDFHIPHACLKDNGAYRCTGYKESCC PVSSNTVKIQVQEPFTRPVLRASSFQPISGNPVTLTCETQLSLERSDVPLRFRFFRDDQTLGLGWSLSPNFQITAMWSKDSGFYWCKAAT MPYSVISDSPRSWIQVQIPASHPVLTLSPEKALNFEGTKVTLHCETQEDSLRTLYRFYHEGVPLRHKSVRCERGASISFSLTTENSGNYY CTADNGLGAKPSKAVSLSVTVPVSHPVLNLSSPEDLIFEGAKVTLHCEAQRGSLPILYQFHHEGAALERRSANSAGGVAISFSLTAEHSG NYYCTADNGFGPQRSKAVSLSVTVPVSHPVLTLSSAEALTFEGATVTLHCEVQRGSPQILYQFYHEDMPLWSSSTPSVGRVSFSFSLTEG HSGNYYCTADNGFGPQRSEVVSLFVTGVFQVAEKMEKRTCALCPKDVEYNVLYFAQSENIAAHENCLLYSSGLVECEDQDPLNPDRSFDV ESVKKEIQRGRKLKCKFCHKRGATVGCDLKNCNKNYHFFCAKKDDAVPQSDGVRGIYKLLCQQHAQFPIIAQSAKFSGVKRKRGRKKPLS GNHVQPPETMKCNTFIRQVKEEHGRHTDATVKVPFLKKCKEAGLLNYLLEEILDKVHSIPEKLMDETTSESDYEEIGSALFDCRLFEDTF VNFQAAIEKKIHASQQRWQQLKEEIELLQDLKQTLCSFQENRDLMSSSTSISSLSY -------------------------------------------------------------- >30034_30034_3_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000368190_PHF11_chr13_50080771_ENST00000378319_length(transcript)=2998nt_BP=1774nt GTGCAGTGTCCTGACTGTAAGATCAAGTCCAAACCTGTTTTGGAATTGAGGAAACTTCTCTTTTGATCTCAGCCCTTGGTGGTCCAGGTC TTCATGCTGCTGTGGGTGATATTACTGGTCCTGGCTCCTGTCAGTGGACAGTTTGCAAGGACACCCAGGCCCATTATTTTCCTCCAGCCT CCATGGACCACAGTCTTCCAAGGAGAGAGAGTGACCCTCACTTGCAAGGGATTTCGCTTCTACTCACCACAGAAAACAAAATGGTACCAT CGGTACCTTGGGAAAGAAATACTAAGAGAAACCCCAGACAATATCCTTGAGGTTCAGGAATCTGGAGAGTACAGATGCCAGGCCCAGGGC TCCCCTCTCAGTAGCCCTGTGCACTTGGATTTTTCTTCAGCTTCGCTGATCCTGCAAGCTCCACTTTCTGTGTTTGAAGGAGACTCTGTG GTTCTGAGGTGCCGGGCAAAGGCGGAAGTAACACTGAATAATACTATTTACAAGAATGATAATGTCCTGGCATTCCTTAATAAAAGAACT GACTTCCATATTCCTCATGCATGTCTCAAGGACAATGGTGCATATCGCTGTACTGGATATAAGGAAAGTTGTTGCCCTGTTTCTTCCAAT ACAGTCAAAATCCAAGTCCAAGAGCCATTTACACGTCCAGTGCTGAGAGCCAGCTCCTTCCAGCCCATCAGCGGGAACCCAGTGACCCTG ACCTGTGAGACCCAGCTCTCTCTAGAGAGGTCAGATGTCCCGCTCCGGTTCCGCTTCTTCAGAGATGACCAGACCCTGGGATTAGGCTGG AGTCTCTCCCCGAATTTCCAGATTACTGCCATGTGGAGTAAAGATTCAGGGTTCTACTGGTGTAAGGCAGCAACAATGCCTTACAGCGTC ATATCTGACAGCCCGAGATCCTGGATACAGGTGCAGATCCCTGCATCTCATCCTGTCCTCACTCTCAGCCCTGAAAAGGCTCTGAATTTT GAGGGAACCAAGGTGACACTTCACTGTGAAACCCAGGAAGATTCTCTGCGCACTTTGTACAGGTTTTATCATGAGGGTGTCCCCCTGAGG CACAAGTCAGTCCGCTGTGAAAGGGGAGCATCCATCAGCTTCTCACTGACTACAGAGAATTCAGGGAACTACTACTGCACAGCTGACAAT GGCCTTGGCGCCAAGCCCAGTAAGGCTGTGAGCCTCTCAGTCACTGTTCCCGTGTCTCATCCTGTCCTCAACCTCAGCTCTCCTGAGGAC CTGATTTTTGAGGGAGCCAAGGTGACACTTCACTGTGAAGCCCAGAGAGGTTCACTCCCCATCCTGTACCAGTTTCATCATGAGGGTGCT GCCCTGGAGCGTAGGTCGGCCAACTCTGCAGGAGGAGTGGCCATCAGCTTCTCTCTGACTGCAGAGCATTCAGGGAACTACTACTGCACA GCTGACAATGGCTTTGGCCCCCAGCGCAGTAAGGCGGTGAGCCTCTCCGTCACTGTCCCTGTGTCTCATCCTGTCCTCACCCTCAGCTCT GCTGAGGCCCTGACTTTTGAAGGAGCCACTGTGACACTTCACTGTGAAGTCCAGAGAGGTTCCCCACAAATCCTATACCAGTTTTATCAT GAGGACATGCCCCTGTGGAGCAGCTCAACACCCTCTGTGGGAAGAGTGTCCTTCAGCTTCTCTCTGACTGAAGGACATTCAGGGAATTAC TACTGCACAGCTGACAATGGCTTTGGTCCCCAGCGCAGTGAAGTGGTGAGCCTTTTTGTCACTGGTGTCTTTCAGGTTGCAGAAAAGATG GAAAAAAGGACATGTGCACTCTGCCCCAAAGATGTCGAATATAATGTCCTATACTTTGCACAATCAGAGAATATAGCTGCTCATGAGAAT TGTTTGCTGTATTCTTCAGGACTTGTGGAATGTGAGGATCAGGATCCACTTAATCCTGATAGAAGTTTTGATGTGGAATCAGTAAAGAAA GAAATCCAGAGAGGAAGGAAGTTGAAATGCAAATTTTGTCATAAAAGAGGAGCCACCGTGGGATGTGATTTAAAAAACTGTAACAAGAAT TACCACTTTTTCTGTGCCAAGAAGGACGACGCAGTTCCACAGTCTGATGGAGTTCGAGGAATTTATAAACTGCTTTGCCAGCAACATGCT CAATTCCCGATCATCGCTCAAAGTGCTAAATTTTCAGGAGTGAAAAGAAAAAGAGGAAGGAAGAAACCCCTCTCAGGCAATCATGTACAG CCACCCGAAACAATGAAATGTAATACATTCATAAGACAAGTGAAAGAAGAGCATGGCAGACACACAGATGCAACTGTGAAAGTTCCTTTT CTTAAGAAATGCAAGGAAGCAGGACTTCTTAATTACTTACTTGAAGAAATATTAGACAAAGTTCATTCAATTCCAGAAAAACTCATGGAT GAGACTACTTCAGAATCAGACTATGAAGAAATCGGGAGTGCACTTTTTGACTGTAGATTGTTCGAAGACACATTTGTAAATTTTCAAGCA GCAATAGAGAAAAAAATTCATGCATCTCAACAAAGGTGGCAGCAGTTGAAGGAAGAGATTGAGCTACTTCAGGACTTAAAACAAACCTTG TGCTCTTTTCAAGAAAATAGAGATCTTATGTCAAGTTCTACATCAATATCATCCCTGTCTTATTAGGGATTACCGTTTCCTAAGCCAAGA GTCATGTCAAATTGCAATCAGGCTCAAAACCAGAGACCAGGCTGTGAAATCCACACATCTTTAGAACTAGTCGTCTCCTCTTGGCCTCAG CAGCTCTTCCCTGTTCTTACTGGTTGACATTTTGATCACTCTTTGCACACTCTTGTGTTTTTTGCTCACTGTCACATTCCCAGCACCTAG TATGCTCAGTAAATGTTTGTGGAATAAGTGCATAAAATGTTCTTAACCTTTGATTCTACTTACAGCCCATGATAGCCTCTTAGATATAAT AAATTTGGATTATACTACTTTACTTGTA >30034_30034_3_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000368190_PHF11_chr13_50080771_ENST00000378319_length(amino acids)=866AA_BP=566 MVVQVFMLLWVILLVLAPVSGQFARTPRPIIFLQPPWTTVFQGERVTLTCKGFRFYSPQKTKWYHRYLGKEILRETPDNILEVQESGEYR CQAQGSPLSSPVHLDFSSASLILQAPLSVFEGDSVVLRCRAKAEVTLNNTIYKNDNVLAFLNKRTDFHIPHACLKDNGAYRCTGYKESCC PVSSNTVKIQVQEPFTRPVLRASSFQPISGNPVTLTCETQLSLERSDVPLRFRFFRDDQTLGLGWSLSPNFQITAMWSKDSGFYWCKAAT MPYSVISDSPRSWIQVQIPASHPVLTLSPEKALNFEGTKVTLHCETQEDSLRTLYRFYHEGVPLRHKSVRCERGASISFSLTTENSGNYY CTADNGLGAKPSKAVSLSVTVPVSHPVLNLSSPEDLIFEGAKVTLHCEAQRGSLPILYQFHHEGAALERRSANSAGGVAISFSLTAEHSG NYYCTADNGFGPQRSKAVSLSVTVPVSHPVLTLSSAEALTFEGATVTLHCEVQRGSPQILYQFYHEDMPLWSSSTPSVGRVSFSFSLTEG HSGNYYCTADNGFGPQRSEVVSLFVTGVFQVAEKMEKRTCALCPKDVEYNVLYFAQSENIAAHENCLLYSSGLVECEDQDPLNPDRSFDV ESVKKEIQRGRKLKCKFCHKRGATVGCDLKNCNKNYHFFCAKKDDAVPQSDGVRGIYKLLCQQHAQFPIIAQSAKFSGVKRKRGRKKPLS GNHVQPPETMKCNTFIRQVKEEHGRHTDATVKVPFLKKCKEAGLLNYLLEEILDKVHSIPEKLMDETTSESDYEEIGSALFDCRLFEDTF VNFQAAIEKKIHASQQRWQQLKEEIELLQDLKQTLCSFQENRDLMSSSTSISSLSY -------------------------------------------------------------- >30034_30034_4_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000368191_PHF11_chr13_50080771_ENST00000378319_length(transcript)=2781nt_BP=1557nt GAGAGCACAGCTTCTCAGATGTGCTCCTTGGAGCTGGTGTGCAGTGTCCTGACTGTAAGATCAAGTCCAAACCTGTTTTGGAATTGAGGA AACTTCTCTTTTGATCTCAGCCCTTGGTGGTCCAGGTCTTCATGCTGCTGTGGGTGATATTACTGGTCCTGGCTCCTGTCAGTGGACAGT TTGCTTCGCTGATCCTGCAAGCTCCACTTTCTGTGTTTGAAGGAGACTCTGTGGTTCTGAGGTGCCGGGCAAAGGCGGAAGTAACACTGA ATAATACTATTTACAAGAATGATAATGTCCTGGCATTCCTTAATAAAAGAACTGACTTCCATATTCCTCATGCATGTCTCAAGGACAATG GTGCATATCGCTGTACTGGATATAAGGAAAGTTGTTGCCCTGTTTCTTCCAATACAGTCAAAATCCAAGTCCAAGAGCCATTTACACGTC CAGTGCTGAGAGCCAGCTCCTTCCAGCCCATCAGCGGGAACCCAGTGACCCTGACCTGTGAGACCCAGCTCTCTCTAGAGAGGTCAGATG TCCCGCTCCGGTTCCGCTTCTTCAGAGATGACCAGACCCTGGGATTAGGCTGGAGTCTCTCCCCGAATTTCCAGATTACTGCCATGTGGA GTAAAGATTCAGGGTTCTACTGGTGTAAGGCAGCAACAATGCCTTACAGCGTCATATCTGACAGCCCGAGATCCTGGATACAGGTGCAGA TCCCTGCATCTCATCCTGTCCTCACTCTCAGCCCTGAAAAGGCTCTGAATTTTGAGGGAACCAAGGTGACACTTCACTGTGAAACCCAGG AAGATTCTCTGCGCACTTTGTACAGGTTTTATCATGAGGGTGTCCCCCTGAGGCACAAGTCAGTCCGCTGTGAAAGGGGAGCATCCATCA GCTTCTCACTGACTACAGAGAATTCAGGGAACTACTACTGCACAGCTGACAATGGCCTTGGCGCCAAGCCCAGTAAGGCTGTGAGCCTCT CAGTCACTGTTCCCGTGTCTCATCCTGTCCTCAACCTCAGCTCTCCTGAGGACCTGATTTTTGAGGGAGCCAAGGTGACACTTCACTGTG AAGCCCAGAGAGGTTCACTCCCCATCCTGTACCAGTTTCATCATGAGGGTGCTGCCCTGGAGCGTAGGTCGGCCAACTCTGCAGGAGGAG TGGCCATCAGCTTCTCTCTGACTGCAGAGCATTCAGGGAACTACTACTGCACAGCTGACAATGGCTTTGGCCCCCAGCGCAGTAAGGCGG TGAGCCTCTCCGTCACTGTCCCTGTGTCTCATCCTGTCCTCACCCTCAGCTCTGCTGAGGCCCTGACTTTTGAAGGAGCCACTGTGACAC TTCACTGTGAAGTCCAGAGAGGTTCCCCACAAATCCTATACCAGTTTTATCATGAGGACATGCCCCTGTGGAGCAGCTCAACACCCTCTG TGGGAAGAGTGTCCTTCAGCTTCTCTCTGACTGAAGGACATTCAGGGAATTACTACTGCACAGCTGACAATGGCTTTGGTCCCCAGCGCA GTGAAGTGGTGAGCCTTTTTGTCACTGGTGTCTTTCAGGTTGCAGAAAAGATGGAAAAAAGGACATGTGCACTCTGCCCCAAAGATGTCG AATATAATGTCCTATACTTTGCACAATCAGAGAATATAGCTGCTCATGAGAATTGTTTGCTGTATTCTTCAGGACTTGTGGAATGTGAGG ATCAGGATCCACTTAATCCTGATAGAAGTTTTGATGTGGAATCAGTAAAGAAAGAAATCCAGAGAGGAAGGAAGTTGAAATGCAAATTTT GTCATAAAAGAGGAGCCACCGTGGGATGTGATTTAAAAAACTGTAACAAGAATTACCACTTTTTCTGTGCCAAGAAGGACGACGCAGTTC CACAGTCTGATGGAGTTCGAGGAATTTATAAACTGCTTTGCCAGCAACATGCTCAATTCCCGATCATCGCTCAAAGTGCTAAATTTTCAG GAGTGAAAAGAAAAAGAGGAAGGAAGAAACCCCTCTCAGGCAATCATGTACAGCCACCCGAAACAATGAAATGTAATACATTCATAAGAC AAGTGAAAGAAGAGCATGGCAGACACACAGATGCAACTGTGAAAGTTCCTTTTCTTAAGAAATGCAAGGAAGCAGGACTTCTTAATTACT TACTTGAAGAAATATTAGACAAAGTTCATTCAATTCCAGAAAAACTCATGGATGAGACTACTTCAGAATCAGACTATGAAGAAATCGGGA GTGCACTTTTTGACTGTAGATTGTTCGAAGACACATTTGTAAATTTTCAAGCAGCAATAGAGAAAAAAATTCATGCATCTCAACAAAGGT GGCAGCAGTTGAAGGAAGAGATTGAGCTACTTCAGGACTTAAAACAAACCTTGTGCTCTTTTCAAGAAAATAGAGATCTTATGTCAAGTT CTACATCAATATCATCCCTGTCTTATTAGGGATTACCGTTTCCTAAGCCAAGAGTCATGTCAAATTGCAATCAGGCTCAAAACCAGAGAC CAGGCTGTGAAATCCACACATCTTTAGAACTAGTCGTCTCCTCTTGGCCTCAGCAGCTCTTCCCTGTTCTTACTGGTTGACATTTTGATC ACTCTTTGCACACTCTTGTGTTTTTTGCTCACTGTCACATTCCCAGCACCTAGTATGCTCAGTAAATGTTTGTGGAATAAGTGCATAAAA TGTTCTTAACCTTTGATTCTACTTACAGCCCATGATAGCCTCTTAGATATAATAAATTTGGATTATACTACTTTACTTGTA >30034_30034_4_FCRL5-PHF11_FCRL5_chr1_157504404_ENST00000368191_PHF11_chr13_50080771_ENST00000378319_length(amino acids)=781AA_BP=481 MVVQVFMLLWVILLVLAPVSGQFASLILQAPLSVFEGDSVVLRCRAKAEVTLNNTIYKNDNVLAFLNKRTDFHIPHACLKDNGAYRCTGY KESCCPVSSNTVKIQVQEPFTRPVLRASSFQPISGNPVTLTCETQLSLERSDVPLRFRFFRDDQTLGLGWSLSPNFQITAMWSKDSGFYW CKAATMPYSVISDSPRSWIQVQIPASHPVLTLSPEKALNFEGTKVTLHCETQEDSLRTLYRFYHEGVPLRHKSVRCERGASISFSLTTEN SGNYYCTADNGLGAKPSKAVSLSVTVPVSHPVLNLSSPEDLIFEGAKVTLHCEAQRGSLPILYQFHHEGAALERRSANSAGGVAISFSLT AEHSGNYYCTADNGFGPQRSKAVSLSVTVPVSHPVLTLSSAEALTFEGATVTLHCEVQRGSPQILYQFYHEDMPLWSSSTPSVGRVSFSF SLTEGHSGNYYCTADNGFGPQRSEVVSLFVTGVFQVAEKMEKRTCALCPKDVEYNVLYFAQSENIAAHENCLLYSSGLVECEDQDPLNPD RSFDVESVKKEIQRGRKLKCKFCHKRGATVGCDLKNCNKNYHFFCAKKDDAVPQSDGVRGIYKLLCQQHAQFPIIAQSAKFSGVKRKRGR KKPLSGNHVQPPETMKCNTFIRQVKEEHGRHTDATVKVPFLKKCKEAGLLNYLLEEILDKVHSIPEKLMDETTSESDYEEIGSALFDCRL FEDTFVNFQAAIEKKIHASQQRWQQLKEEIELLQDLKQTLCSFQENRDLMSSSTSISSLSY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FCRL5-PHF11 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FCRL5-PHF11 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FCRL5-PHF11 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
