
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CIRH1A-ZFPM1 (FusionGDB2 ID:HG84916TG161882) |
Fusion Gene Summary for CIRH1A-ZFPM1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CIRH1A-ZFPM1 | Fusion gene ID: hg84916tg161882 | Hgene | Tgene | Gene symbol | CIRH1A | ZFPM1 | Gene ID | 84916 | 161882 |
| Gene name | UTP4 small subunit processome component | zinc finger protein, FOG family member 1 | |
| Synonyms | CIRH1A|CIRHIN|NAIC|TEX292 | FOG|FOG1|ZC2HC11A|ZNF408|ZNF89A | |
| Cytomap | ('CIRH1A')('ZFPM1') 16q22.1 | 16q24.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | U3 small nucleolar RNA-associated protein 4 homologUTP4 small subunit (SSU) processome componentUTP4, small subunit (SSU) processome component, homologcirrhosis, autosomal recessive 1A (cirhin)testis expressed gene 292 | zinc finger protein ZFPM1FOG-1friend of GATA 1friend of GATA protein 1zinc finger protein 89Azinc finger protein, multitype 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000563094, ENST00000314423, ENST00000352319, ENST00000569615, | ||
| Fusion gene scores | * DoF score | 4 X 5 X 3=60 | 6 X 5 X 3=90 |
| # samples | 5 | 6 | |
| ** MAII score | log2(5/60*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CIRH1A [Title/Abstract] AND ZFPM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CIRH1A(69201088)-ZFPM1(88593221), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CIRH1A-ZFPM1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CIRH1A-ZFPM1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CIRH1A | GO:0006355 | regulation of transcription, DNA-templated | 19732766 |
| Tgene | ZFPM1 | GO:0010724 | regulation of definitive erythrocyte differentiation | 15920471 |
| Tgene | ZFPM1 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process | 21646796 |
| Tgene | ZFPM1 | GO:0045403 | negative regulation of interleukin-4 biosynthetic process | 21646796 |
 Fusion gene breakpoints across CIRH1A (5'-gene) Fusion gene breakpoints across CIRH1A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
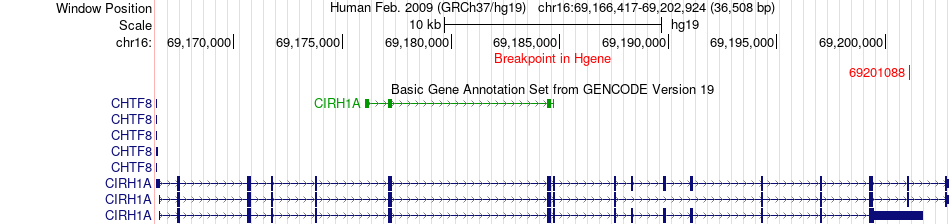 |
 Fusion gene breakpoints across ZFPM1 (3'-gene) Fusion gene breakpoints across ZFPM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
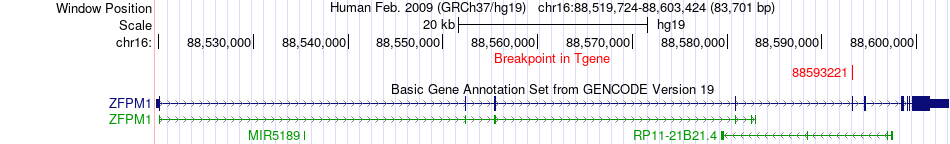 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8588 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
Top |
Fusion Gene ORF analysis for CIRH1A-ZFPM1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000563094 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| 3UTR-intron | ENST00000563094 | ENST00000569086 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| 5CDS-intron | ENST00000314423 | ENST00000569086 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| 5CDS-intron | ENST00000352319 | ENST00000569086 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| In-frame | ENST00000314423 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| In-frame | ENST00000352319 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| intron-3CDS | ENST00000569615 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
| intron-intron | ENST00000569615 | ENST00000569086 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000314423 | CIRH1A | chr16 | 69201088 | + | ENST00000319555 | ZFPM1 | chr16 | 88593221 | + | 6777 | 2121 | 54 | 4739 | 1561 |
| ENST00000352319 | CIRH1A | chr16 | 69201088 | + | ENST00000319555 | ZFPM1 | chr16 | 88593221 | + | 6297 | 1641 | 42 | 4259 | 1405 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000314423 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + | 0.004180356 | 0.99581957 |
| ENST00000352319 | ENST00000319555 | CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + | 0.003301543 | 0.99669844 |
Top |
Fusion Genomic Features for CIRH1A-ZFPM1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + | 0.001462965 | 0.99853706 |
| CIRH1A | chr16 | 69201088 | + | ZFPM1 | chr16 | 88593221 | + | 0.001462965 | 0.99853706 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
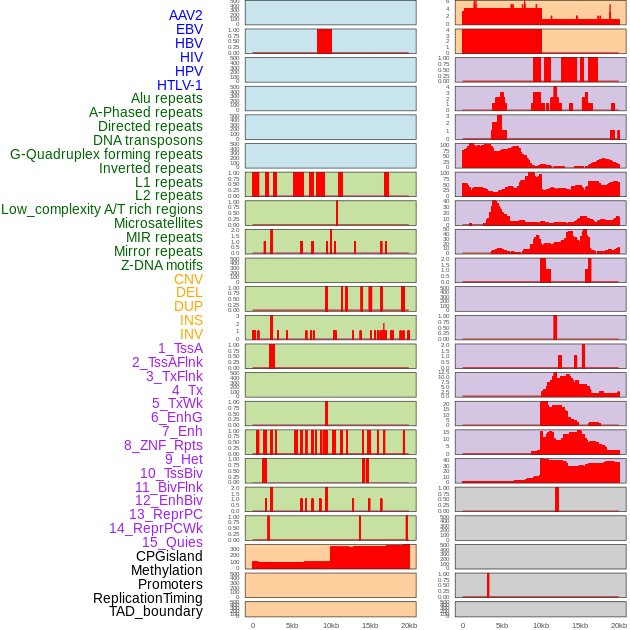 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
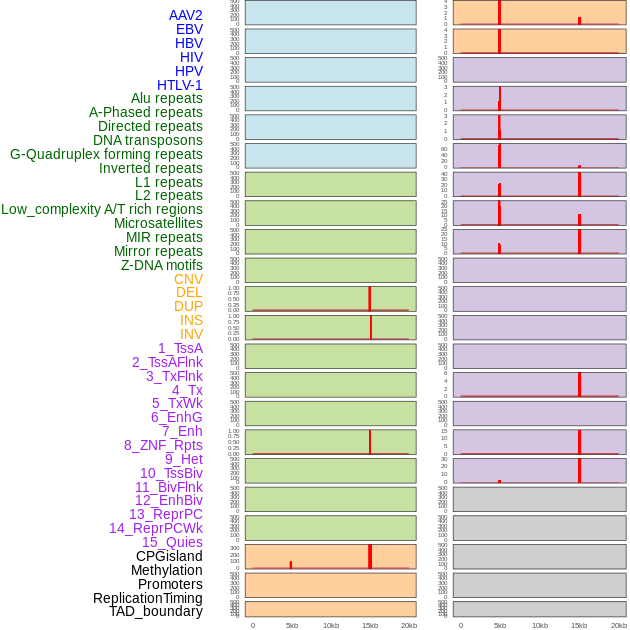 |
Top |
Fusion Protein Features for CIRH1A-ZFPM1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:69201088/chr16:88593221) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 136_181 | 648 | 687.0 | Repeat | Note=WD 4 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 182_226 | 648 | 687.0 | Repeat | Note=WD 5 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 227_275 | 648 | 687.0 | Repeat | Note=WD 6 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 276_317 | 648 | 687.0 | Repeat | Note=WD 7 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 318_377 | 648 | 687.0 | Repeat | Note=WD 8 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 378_427 | 648 | 687.0 | Repeat | Note=WD 9 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 428_475 | 648 | 687.0 | Repeat | Note=WD 10 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 476_516 | 648 | 687.0 | Repeat | Note=WD 11 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 517_566 | 648 | 687.0 | Repeat | Note=WD 12 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 51_92 | 648 | 687.0 | Repeat | Note=WD 2 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 567_627 | 648 | 687.0 | Repeat | Note=WD 13 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 7_50 | 648 | 687.0 | Repeat | Note=WD 1 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 93_135 | 648 | 687.0 | Repeat | Note=WD 3 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 136_181 | 533 | 572.0 | Repeat | Note=WD 4 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 182_226 | 533 | 572.0 | Repeat | Note=WD 5 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 227_275 | 533 | 572.0 | Repeat | Note=WD 6 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 276_317 | 533 | 572.0 | Repeat | Note=WD 7 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 318_377 | 533 | 572.0 | Repeat | Note=WD 8 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 378_427 | 533 | 572.0 | Repeat | Note=WD 9 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 428_475 | 533 | 572.0 | Repeat | Note=WD 10 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 476_516 | 533 | 572.0 | Repeat | Note=WD 11 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 51_92 | 533 | 572.0 | Repeat | Note=WD 2 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 7_50 | 533 | 572.0 | Repeat | Note=WD 1 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 93_135 | 533 | 572.0 | Repeat | Note=WD 3 |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 235_268 | 134 | 1007.0 | Zinc finger | CCHC FOG-type 1 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 290_314 | 134 | 1007.0 | Zinc finger | C2H2-type 1 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 320_342 | 134 | 1007.0 | Zinc finger | C2H2-type 2 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 348_371 | 134 | 1007.0 | Zinc finger | C2H2-type 3 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 571_604 | 134 | 1007.0 | Zinc finger | CCHC FOG-type 2 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 677_710 | 134 | 1007.0 | Zinc finger | CCHC FOG-type 3 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 811_844 | 134 | 1007.0 | Zinc finger | CCHC FOG-type 4 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 854_877 | 134 | 1007.0 | Zinc finger | C2H2-type 4 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 968_1001 | 134 | 1007.0 | Zinc finger | CCHC FOG-type 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000314423 | + | 16 | 17 | 628_666 | 648 | 687.0 | Repeat | Note=WD 14 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 517_566 | 533 | 572.0 | Repeat | Note=WD 12 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 567_627 | 533 | 572.0 | Repeat | Note=WD 13 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000352319 | + | 13 | 14 | 628_666 | 533 | 572.0 | Repeat | Note=WD 14 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 136_181 | 0 | 626.0 | Repeat | Note=WD 4 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 182_226 | 0 | 626.0 | Repeat | Note=WD 5 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 227_275 | 0 | 626.0 | Repeat | Note=WD 6 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 276_317 | 0 | 626.0 | Repeat | Note=WD 7 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 318_377 | 0 | 626.0 | Repeat | Note=WD 8 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 378_427 | 0 | 626.0 | Repeat | Note=WD 9 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 428_475 | 0 | 626.0 | Repeat | Note=WD 10 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 476_516 | 0 | 626.0 | Repeat | Note=WD 11 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 517_566 | 0 | 626.0 | Repeat | Note=WD 12 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 51_92 | 0 | 626.0 | Repeat | Note=WD 2 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 567_627 | 0 | 626.0 | Repeat | Note=WD 13 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 628_666 | 0 | 626.0 | Repeat | Note=WD 14 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 7_50 | 0 | 626.0 | Repeat | Note=WD 1 |
| Hgene | CIRH1A | chr16:69201088 | chr16:88593221 | ENST00000563094 | + | 1 | 15 | 93_135 | 0 | 626.0 | Repeat | Note=WD 3 |
Top |
Fusion Gene Sequence for CIRH1A-ZFPM1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >16843_16843_1_CIRH1A-ZFPM1_CIRH1A_chr16_69201088_ENST00000314423_ZFPM1_chr16_88593221_ENST00000319555_length(transcript)=6777nt_BP=2121nt GCGGCCAACGGGCGACAACCGAACCTCCCGCCGCCGTCGCCGCCGCCGCGAGCACTGCCTGCGCACTTCCGACTATGCGCCAGGAGCACT TCCGGGGCAGAGCCTGAGAGCGCGCGCGCACGTGGGGCCGGGGCGGAGAGAGGCGAGCACCGGGAAGGGGAGCGTGGGGCCGCTGGAATG GGTGAATTTAAGGTCCATCGAGTACGTTTCTTTAATTATGTTCCATCAGGAATCCGCTGTGTGGCTTACAATAACCAGTCAAACAGATTG GCTGTTTCACGAACAGATGGCACTGTGGAAATTTATAACTTGTCAGCAAACTACTTTCAGGAGAAATTTTTCCCAGGTCATGAGTCTCGG GCTACAGAAGCTTTGTGCTGGGCAGAAGGACAGCGACTCTTTAGTGCTGGGCTCAATGGCGAGATTATGGAGTATGATTTACAGGCGTTA AACATCAAGTATGCTATGGATGCCTTTGGAGGACCTATTTGGAGCATGGCTGCCAGCCCCAGTGGCTCTCAACTTTTGGTTGGTTGTGAA GATGGATCTGTGAAACTATTTCAAATTACCCCAGACAAAATCCAGTTTGAAAGAAATTTTGATCGGCAGAAAAGTCGCATCCTGAGTCTC AGCTGGCATCCCTCTGGTACCCACATTGCAGCTGGTTCCATAGACTACATTAGTGTGTTTGATGTCAAATCAGGCAGCGCTGTTCATAAG ATGATTGTGGACAGGCAGTATATGGGCGTGTCTAAGCGGAAGTGCATCGTGTGGGGTGTCGCCTTCTTGTCCGATGGCACTATCATAAGT GTGGACTCTGCTGGGAAGGTGCAGTTCTGGGACTCAGCCACTGGGACGCTTGTGAAGAGCCATCTCATCGCTAATGCTGACGTGCAGTCC ATTGCTGTAGCTGACCAAGAAGACAGTTTCGTGGTGGGCACAGCCGAGGGAACAGTCTTCCATTTTCAGCTGGTCCCTGTGACATCTAAC AGCAGTGAGAAGCAGTGGGTGCGGACAAAACCGTTCCAGCATCACACTCATGACGTGCGCACTGTGGCCCACAGCCCAACAGCGCTGATA TCTGGAGGCACTGACACCCACTTAGTCTTTCGTCCTCTCATGGAGAAGGTGGAAGTAAAGAATTACGATGCCGCTCTCCGAAAAATCACC TTTCCCCACCGATGTCTCATCTCCTGTTCTAAAAAGAGGCAGCTTCTCCTCTTCCAGTTTGCTCATCACTTAGAACTTTGGCGACTGGGA TCCACAGTTGCAACAGGCAAGAATGGGGATACTCTTCCACTCTCTAAAAATGCAGATCATTTACTGCACCTAAAGACAAAGGGTCCTGAG AACATTATCTGTAGCTGTATCTCCCCATGTGGAAGTTGGATAGCCTATTCTACAGTTTCTCGGTTTTTTCTCTATCGGCTGAATTATGAA CATGACAACATAAGCCTCAAAAGGGTTTCCAAAATGCCAGCATTCCTTCGCTCTGCCCTTCAGATTTTGTTTTCTGAAGATTCAACAAAG CTCTTTGTAGCATCAAATCAAGGAGCTCTGCATATTGTTCAGCTGTCAGGAGGAAGCTTCAAGCACCTGCATGCTTTCCAGCCTCAGTCA GGAACAGTGGAGGCCATGTGTCTTTTGGCAGTCAGTCCAGATGGGAATTGGCTAGCTGCATCAGGTACCAGTGCTGGAGTCCATGTCTAC AACGTAAAACAGCTAAAGCTTCACTGCACGGTGCCTGCTTACAATTTCCCAGTGACTGCTATGGCTATTGCCCCCAATACCAACAACCTT GTCATCGCTCATTCGGACCAGCAGGTATTTGAGTACAGCATCCCAGACAAACAGTATACAGATTGGAGCCGGACTGTCCAGAAGCAGGGC TTTCACCACCTTTGGCTCCAAAGGGATACTCCTATCACACACATCAGTTTTCATCCCAAGAGACCGATGCACATCCTTCTCCATGATGCC TACATGTTCTGCATCATTGACAAGTCATTGCCCCTTCCAAATGACAAAACCTTACTCTACAATCCATTTCCTCCCACGAATGAATCAGAT GTCATCCGGAGGCGCACAGCTCATGCTTTTAAAATTTCTAAGATATATAAGAGCCCAGCCCTGACCCTGCTGCTGGTGGACGAGGCCTGC TGGCTGAGGACGCTGCCCCAGGCCCTGACTGAGGCCGAGGCCAACACAGAGATCCACAGGAAGGATGACGCACTCTGGTGCAGGGTCACC AAGCCGGTGCCTGCGGGGGGACTCCTGAGCGTGCTCCTCACGGCCGAGCCCCACAGCACCCCCGGCCACCCTGTGAAGAAGGAGCCAGCA GAGCCCACGTGCCCGGCCCCTGCACACGACCTCCAGCTCCTGCCCCAGCAGGCCGGGATGGCCTCCATCCTTGCCACCGCAGTGATCAAC AAAGACGTCTTCCCCTGCAAGGACTGTGGCATCTGGTACCGCAGCGAGCGCAACCTGCAGGCGCACCTGCTCTACTACTGCGCCAGCCGC CAGGGCACCGGCTCCCCGGCCGCAGCCGCCACAGACGAGAAGCCCAAAGAGACCTACCCCAACGAGCGCGTCTGCCCCTTCCCCCAGTGC CGCAAGAGCTGCCCCAGCGCCAGCTCCCTGGAGATCCACATGCGCAGCCACAGCGGAGAGCGGCCCTTCGTGTGCCTGATCTGCCTGTCG GCCTTCACCACCAAGGCCAACTGCGAGCGGCACCTCAAGGTGCACACGGACACGCTGAGCGGTGTCTGCCACAGCTGTGGCTTCATCTCC ACCACAAGGGACATCCTCTACAGCCACTTGGTCACCAACCACATGGTCTGCCAGCCTGGCTCCAAGGGTGAGATCTACTCGCCAGGGGCC GGACACCCAGCAACCAAGCTGCCCCCAGACAGTCTGGGCAGCTTCCAGCAGCAGCACACGGCCCTGCAAGGCCCCCTGGCCTCCGCGGAC CTGGGCCTGGCGCCCACCCCATCGCCAGGACTGGACAGAAAGGCCCTGGCCGAGGCCACCAACGGAGAGGCCAGAGCGGAGCCTCTGGCC CAGAATGGAGGCAGCAGCGAGCCCCCGGCGGCCCCCAGGAGCATCAAGGTGGAGGCGGTGGAGGAGCCGGAGGCGGCCCCCATCCTGGGC CCCGGAGAGCCTGGGCCCCAGGCCCCGTCGCGGACGCCGTCGCCGCGCAGCCCCGCCCCGGCCAGGGTCAAGGCCGAGCTGTCCAGCCCC ACGCCGGGCTCCAGCCCGGTGCCCGGCGAGCTGGGCCTGGCCGGGGCCCTGTTCCTTCCGCAGTACGTGTTCGGGCCCGACGCGGCGCCC CCCGCCTCGGAGATCCTGGCCAAGATGTCCGAGCTGGTGCACAGCCGGCTGCAGCAGGGCGCGGGCGCGGGCGCCGGCGGCGCGCAGACC GGGCTCTTCCCCGGGGCCCCCAAGGGCGCTACGTGCTTCGAGTGCGAGATCACCTTCAGCAACGTCAACAACTACTACGTGCACAAGCGC CTCTACTGTTCAGGCCGCCGTGCGCCCGAGGACGCGCCTGCCGCGCGCAGGCCCAAGGCGCCCCCCGGCCCGGCCCGCGCGCCCCCCGGC CAGCCCGCCGAACCCGACGCGCCGCGCTCGTCCCCGGGCCCCGGAGCGCGCGAGGAGGGGGCTGGGGGCGCGGCCACGCCCGAGGACGGC GCGGGCGGCCGGGGCAGCGAGGGCAGCCAGAGCCCGGGTAGCTCCGTGGACGACGCGGAGGACGACCCCAGCCGCACGCTGTGCGAGGCC TGCAACATCCGCTTCAGCCGCCACGAGACCTACACCGTGCACAAGCGGTACTACTGCGCCTCGCGCCACGACCCGCCGCCGCGCCGACCG GCCGCGCCCCCGGGACCCCCTGGGCCGGCCGCGCCCCCGGCCCCCTCTCCCGCCGCGCCTGTGCGCACGCGCAGACGCCGCAAGCTCTAC GAGCTGCACGCGGCCGGCGCCCCGCCCCCCCCGCCGCCCGGCCACGCCCCCGCGCCCGAGTCGCCGCGGCCCGGAAGCGGAAGCGGAAGC GGCCCCGGCCTCGCCCCTGCGCGCTCGCCCGGCCCCGCGGCCGACGGCCCCATCGACCTGAGCAAGAAGCCGCGGCGCCCGCTCCCCGGA GCCCCGGCACCGGCGCTGGCCGACTACCACGAGTGCACGGCCTGCCGCGTGAGCTTCCACAGCCTCGAGGCCTACCTGGCGCACAAGAAG TACTCGTGCCCCGCTGCGCCACCGCCCGGCGCGCTCGGCCTGCCCGCCGCCGCCTGCCCCTACTGCCCCCCGAACGGCCCGGTGCGCGGG GACCTGCTGGAGCATTTCCGCCTGGCGCACGGCCTGCTGCTCGGCGCGCCCCTGGCCGGCCCGGGGGTCGAGGCCCGGACGCCGGCCGAC CGCGGCCCCTCGCCCGCTCCCGCCCCCGCCGCCTCCCCGCAGCCCGGGTCCCGTGGCCCCCGGGACGGCCTCGGCCCGGAGCCCCAGGAG CCGCCGCCCGGCCCGCCCCCGTCCCCGGCCGCCGCGCCCGAGGCCGTGCCGCCCCCGCCGGCGCCCCCCTCCTACTCGGACAAGGGCGTC CAGACTCCCAGCAAGGGCACGCCGGCGCCGCTGCCCAACGGCAACCACCGGTACTGCCGTCTTTGCAACATCAAGTTCAGCAGCCTGTCC ACCTTCATCGCCCACAAGAAGTATTACTGCTCCTCGCACGCCGCCGAGCACGTGAAGTGAGCGCCCACACTACAGCCGCAGACGCTTTGC ACGCCCCGCTGCGATGCGGGGAGGGGGCCGCCCCCAGGCCGCACGGACTGCCGCTCCTGGGAACCCCGCCACGCACAGGCCTCGGCGGAG GGGGCCGCAGGGGGCAGCGCCCGCCTGGACCCTTGGCACTTAATAAAGAAGTTCAGTTTGATGAGCATGGTGGTGGGCCAGCCTAGTTCT CTGAGCCAGCAGGCACACGCAGCCAGTGTCACATCCAGCCCTGCTGTGTACACAGGCCACTGCGGCCCGGAGTGGCTGGGCCCCTGCCGG AGTTACACCACTGGGTGCTAAGAGCTAGACCGAGCGTCAGGTCGGAGGCCACAGCACACACTGAACCACTGCAGGGCCAGCCGCTCTGGG CCTACCCCACCCCAGCCCTGTCCATACCCCCCTAGGGAGAGCAGCCTCCACACTGCTGACCCTCTCCACGGTCTACTTGGCCGCCAGACA CGGAAGAGAGGGGCACTGACCTGGCTGGCCATCCCCCAACCCTGCATGGTCCTGGGGGTCCGATGTCCCCCATCCAGTCACAGACCACCA GGGATGGCAGGGGTGGGGGCGCCCTGGGGAGGATGAAGCCCTGTGGTGGCCAAGGCTGGGGGAAGAACAGGCGAGTAGGTGGACAGGAGA GGGAGACCGAGGCCGAAGCATTCCCTGGCCTCGAGGGGGTCCTGATGCTCACTGGTGGGTGCATCTCTCCCTCAGAGCCAGGGCTGAGGA CAACCGTGTGGTGGAGTCACCTGTGGTGGCTGAGTCGGTGCAAGGGTCAGGGCACCCCCACCCCCCGTCTTCCCTGCCCTGCTGGGCTGT ATGGGCCGGCCACACTGAGGGAGTACTTGGCTGACCGCGTTCAGCCACGGTGTCACCGTGCCGTTTGCAGCTGGGTGGCTGTGGGCAGCC TCCAGCGTGCTTTCTCACTGGAGAACAAAAACACTCTTCTATTCCCAACGTCACTTTATTTTTTTATTTTTGGGTCTCACTCTCTTGCCC AGGCTGGAGTCCCTGGGATACTCACGGCTCATTGCAGCCTTGACCTCGTGAGCTCAAGTGATCTTCCCACCTCAGTTTCCTCAGTAGCTG GGGCTACAGGTGTACACCACCATGCCCAGCTAGTTTTTGTGTTTTTTGTAGAAATGGTGTCTCCCTATGTTGCCCAGGCTGGGCAACCCT ATATTTTTTAGAGCTAAGGTCTTGTTCGGTTCCCCAGGCTGAAGTTCAGTGGCACGATCATAAGTCACTGCAGCCTCAAACTCCTGAGCT GAAACCATCTTCCTGCCTCAGCCTCCCAAGTAGCTGGGACTACAGGCTTGACCCACCACACCTGGCTAACTTTTAAAACATTTTTATAGA GACAGGGTCTTCCTGTGTTGCCCAAGATGGTCTCAAACTCCTGACCTCCAGTGCTCCTGCAGCCTCAGCCTCCCAGAACATTAGGATGAC AGGCGCGAGCTACCATTCCCAGCCCATTTCCAACTCTACTTGAACTAGCTGCATTTTTTTCAGCTTTGCCCAGCAGCCCTGGTGAGTGCA GGCAAAGAAGGCAGCGGCCTCTTGCCTCAGAAGCCCGAGGCACTTCCCAGCCAGTCCAGGGCTCGCTCTCCTGCCCGCAGCTCGTGGGCC TGGGTCTGCAGCGCTTTCAGTGGTCTCTGATGTCTTTGTCCACCTGCCCACTAGGGTCTGGCCAGATGGGGCCCGTAGCCAAGGCCAATG GACAGACATGGCTTCCACCCCCTCCCAGGGACCTGAGCTCCAGGGAAGGCAAGCAGCCTTTTCCTCACAGACACCCCTTCTGTTGAAGAC CAGGTGATAGCGGGGACGGGAGGCACGGACCCCTCAAGTCTGCTCATCATTTGCATTGTTGTGCATGAATTTGCGTGGAGGGTTCAGGAG GCTCTGCCCCCAAGTCCTCCCCAGGTGCGGGCGCAGGGTACAAGATGTTCTAAAAAATAGAAACCTGGTGGGGGGTTGTCACTGTGTGTA CATTTAAAAGTAAACTGTTTAAAGGTG >16843_16843_1_CIRH1A-ZFPM1_CIRH1A_chr16_69201088_ENST00000314423_ZFPM1_chr16_88593221_ENST00000319555_length(amino acids)=1561AA_BP=1191 MPAHFRLCARSTSGAEPESARARGAGAERGEHREGERGAAGMGEFKVHRVRFFNYVPSGIRCVAYNNQSNRLAVSRTDGTVEIYNLSANY FQEKFFPGHESRATEALCWAEGQRLFSAGLNGEIMEYDLQALNIKYAMDAFGGPIWSMAASPSGSQLLVGCEDGSVKLFQITPDKIQFER NFDRQKSRILSLSWHPSGTHIAAGSIDYISVFDVKSGSAVHKMIVDRQYMGVSKRKCIVWGVAFLSDGTIISVDSAGKVQFWDSATGTLV KSHLIANADVQSIAVADQEDSFVVGTAEGTVFHFQLVPVTSNSSEKQWVRTKPFQHHTHDVRTVAHSPTALISGGTDTHLVFRPLMEKVE VKNYDAALRKITFPHRCLISCSKKRQLLLFQFAHHLELWRLGSTVATGKNGDTLPLSKNADHLLHLKTKGPENIICSCISPCGSWIAYST VSRFFLYRLNYEHDNISLKRVSKMPAFLRSALQILFSEDSTKLFVASNQGALHIVQLSGGSFKHLHAFQPQSGTVEAMCLLAVSPDGNWL AASGTSAGVHVYNVKQLKLHCTVPAYNFPVTAMAIAPNTNNLVIAHSDQQVFEYSIPDKQYTDWSRTVQKQGFHHLWLQRDTPITHISFH PKRPMHILLHDAYMFCIIDKSLPLPNDKTLLYNPFPPTNESDVIRRRTAHAFKISKIYKSPALTLLLVDEACWLRTLPQALTEAEANTEI HRKDDALWCRVTKPVPAGGLLSVLLTAEPHSTPGHPVKKEPAEPTCPAPAHDLQLLPQQAGMASILATAVINKDVFPCKDCGIWYRSERN LQAHLLYYCASRQGTGSPAAAATDEKPKETYPNERVCPFPQCRKSCPSASSLEIHMRSHSGERPFVCLICLSAFTTKANCERHLKVHTDT LSGVCHSCGFISTTRDILYSHLVTNHMVCQPGSKGEIYSPGAGHPATKLPPDSLGSFQQQHTALQGPLASADLGLAPTPSPGLDRKALAE ATNGEARAEPLAQNGGSSEPPAAPRSIKVEAVEEPEAAPILGPGEPGPQAPSRTPSPRSPAPARVKAELSSPTPGSSPVPGELGLAGALF LPQYVFGPDAAPPASEILAKMSELVHSRLQQGAGAGAGGAQTGLFPGAPKGATCFECEITFSNVNNYYVHKRLYCSGRRAPEDAPAARRP KAPPGPARAPPGQPAEPDAPRSSPGPGAREEGAGGAATPEDGAGGRGSEGSQSPGSSVDDAEDDPSRTLCEACNIRFSRHETYTVHKRYY CASRHDPPPRRPAAPPGPPGPAAPPAPSPAAPVRTRRRRKLYELHAAGAPPPPPPGHAPAPESPRPGSGSGSGPGLAPARSPGPAADGPI DLSKKPRRPLPGAPAPALADYHECTACRVSFHSLEAYLAHKKYSCPAAPPPGALGLPAAACPYCPPNGPVRGDLLEHFRLAHGLLLGAPL AGPGVEARTPADRGPSPAPAPAASPQPGSRGPRDGLGPEPQEPPPGPPPSPAAAPEAVPPPPAPPSYSDKGVQTPSKGTPAPLPNGNHRY CRLCNIKFSSLSTFIAHKKYYCSSHAAEHVK -------------------------------------------------------------- >16843_16843_2_CIRH1A-ZFPM1_CIRH1A_chr16_69201088_ENST00000352319_ZFPM1_chr16_88593221_ENST00000319555_length(transcript)=6297nt_BP=1641nt GAGAGAGGCGAGCACCGGGAAGGGGAGCGTGGGGCCGCTGGAATGGGTGAATTTAAGGTCCATCGAGTACGTTTCTTTAATTATGTTCCA TCAGGAATCCGCTGTGTGGCTTACAATAACCAGTCAAACAGATTGGCTGTTTCACGAACAGATGGCACTGTGGAAATTTATAACTTGTCA GCAAACTACTTTCAGGAGAAATTTTTCCCAGGTCATGAGTCTCGGGCTACAGAAGCTTTGTGCTGGGCAGAAGGACAGCGACTCTTTAGT GCTGGGCTCAATGGCGAGATTATGGAGTATGATTTACAGGCGTTAAACATCAAGTATGCTATGGATGCCTTTGGAGGACCTATTTGGAGC ATGGCTGCCAGCCCCAGTGGCTCTCAACTTTTGGTTGGTTGTGAAGATGGATCTGTGAAACTATTTCAAATTACCCCAGACAAAATCCAG TTTGAAAGAAATTTTGATCGGCAGAAAAGTCGCATCCTGAGTCTCAGCTGGCATCCCTCTGGTACCCACATTGCAGCTGGTTCCATAGAC TACATTAGTGTGTTTGATGTCAAATCAGGCAGCGCTGTTCATAAGATGATTGTGGACAGGCAGTATATGGGCGTGTCTAAGCGGAAGTGC ATCGTGTGGGGTGTCGCCTTCTTGTCCGATGGCACTATCATAAGTGTGGACTCTGCTGGGAAGGTGCAGTTCTGGGACTCAGCCACTGGG ACGCTTGTGAAGAGCCATCTCATCGCTAATGCTGACGTGCAGTCCATTGCTGTAGCTGACCAAGAAGACAGTTTCGTGGTGGGCACAGCC GAGGGAACAGTCTTCCATTTTCAGCTGGTCCCTGTGACATCTAACAGCAGTGAGAAGCAGTGGGTGCGGACAAAACCGTTCCAGCATCAC ACTCATGACGTGCGCACTGTGGCCCACAGCCCAACAGCGCTGATATCTGGAGGCACTGACACCCACTTAGTCTTTCGTCCTCTCATGGAG AAGGTGGAAGTAAAGAATTACGATGCCGCTCTCCGAAAAATCACCTTTCCCCACCGATGTCTCATCTCCTGTTCTAAAAAGAGGCAGCTT CTCCTCTTCCAGTTTGCTCATCACTTAGAACTTTGGCGACTGGGATCCACAGTTGCAACAGGAACAGTGGAGGCCATGTGTCTTTTGGCA GTCAGTCCAGATGGGAATTGGCTAGCTGCATCAGGTACCAGTGCTGGAGTCCATGTCTACAACGTAAAACAGCTAAAGCTTCACTGCACG GTGCCTGCTTACAATTTCCCAGTGACTGCTATGGCTATTGCCCCCAATACCAACAACCTTGTCATCGCTCATTCGGACCAGCAGGTATTT GAGTACAGCATCCCAGACAAACAGTATACAGATTGGAGCCGGACTGTCCAGAAGCAGGGCTTTCACCACCTTTGGCTCCAAAGGGATACT CCTATCACACACATCAGTTTTCATCCCAAGAGACCGATGCACATCCTTCTCCATGATGCCTACATGTTCTGCATCATTGACAAGTCATTG CCCCTTCCAAATGACAAAACCTTACTCTACAATCCATTTCCTCCCACGAATGAATCAGATGTCATCCGGAGGCGCACAGCTCATGCTTTT AAAATTTCTAAGATATATAAGAGCCCAGCCCTGACCCTGCTGCTGGTGGACGAGGCCTGCTGGCTGAGGACGCTGCCCCAGGCCCTGACT GAGGCCGAGGCCAACACAGAGATCCACAGGAAGGATGACGCACTCTGGTGCAGGGTCACCAAGCCGGTGCCTGCGGGGGGACTCCTGAGC GTGCTCCTCACGGCCGAGCCCCACAGCACCCCCGGCCACCCTGTGAAGAAGGAGCCAGCAGAGCCCACGTGCCCGGCCCCTGCACACGAC CTCCAGCTCCTGCCCCAGCAGGCCGGGATGGCCTCCATCCTTGCCACCGCAGTGATCAACAAAGACGTCTTCCCCTGCAAGGACTGTGGC ATCTGGTACCGCAGCGAGCGCAACCTGCAGGCGCACCTGCTCTACTACTGCGCCAGCCGCCAGGGCACCGGCTCCCCGGCCGCAGCCGCC ACAGACGAGAAGCCCAAAGAGACCTACCCCAACGAGCGCGTCTGCCCCTTCCCCCAGTGCCGCAAGAGCTGCCCCAGCGCCAGCTCCCTG GAGATCCACATGCGCAGCCACAGCGGAGAGCGGCCCTTCGTGTGCCTGATCTGCCTGTCGGCCTTCACCACCAAGGCCAACTGCGAGCGG CACCTCAAGGTGCACACGGACACGCTGAGCGGTGTCTGCCACAGCTGTGGCTTCATCTCCACCACAAGGGACATCCTCTACAGCCACTTG GTCACCAACCACATGGTCTGCCAGCCTGGCTCCAAGGGTGAGATCTACTCGCCAGGGGCCGGACACCCAGCAACCAAGCTGCCCCCAGAC AGTCTGGGCAGCTTCCAGCAGCAGCACACGGCCCTGCAAGGCCCCCTGGCCTCCGCGGACCTGGGCCTGGCGCCCACCCCATCGCCAGGA CTGGACAGAAAGGCCCTGGCCGAGGCCACCAACGGAGAGGCCAGAGCGGAGCCTCTGGCCCAGAATGGAGGCAGCAGCGAGCCCCCGGCG GCCCCCAGGAGCATCAAGGTGGAGGCGGTGGAGGAGCCGGAGGCGGCCCCCATCCTGGGCCCCGGAGAGCCTGGGCCCCAGGCCCCGTCG CGGACGCCGTCGCCGCGCAGCCCCGCCCCGGCCAGGGTCAAGGCCGAGCTGTCCAGCCCCACGCCGGGCTCCAGCCCGGTGCCCGGCGAG CTGGGCCTGGCCGGGGCCCTGTTCCTTCCGCAGTACGTGTTCGGGCCCGACGCGGCGCCCCCCGCCTCGGAGATCCTGGCCAAGATGTCC GAGCTGGTGCACAGCCGGCTGCAGCAGGGCGCGGGCGCGGGCGCCGGCGGCGCGCAGACCGGGCTCTTCCCCGGGGCCCCCAAGGGCGCT ACGTGCTTCGAGTGCGAGATCACCTTCAGCAACGTCAACAACTACTACGTGCACAAGCGCCTCTACTGTTCAGGCCGCCGTGCGCCCGAG GACGCGCCTGCCGCGCGCAGGCCCAAGGCGCCCCCCGGCCCGGCCCGCGCGCCCCCCGGCCAGCCCGCCGAACCCGACGCGCCGCGCTCG TCCCCGGGCCCCGGAGCGCGCGAGGAGGGGGCTGGGGGCGCGGCCACGCCCGAGGACGGCGCGGGCGGCCGGGGCAGCGAGGGCAGCCAG AGCCCGGGTAGCTCCGTGGACGACGCGGAGGACGACCCCAGCCGCACGCTGTGCGAGGCCTGCAACATCCGCTTCAGCCGCCACGAGACC TACACCGTGCACAAGCGGTACTACTGCGCCTCGCGCCACGACCCGCCGCCGCGCCGACCGGCCGCGCCCCCGGGACCCCCTGGGCCGGCC GCGCCCCCGGCCCCCTCTCCCGCCGCGCCTGTGCGCACGCGCAGACGCCGCAAGCTCTACGAGCTGCACGCGGCCGGCGCCCCGCCCCCC CCGCCGCCCGGCCACGCCCCCGCGCCCGAGTCGCCGCGGCCCGGAAGCGGAAGCGGAAGCGGCCCCGGCCTCGCCCCTGCGCGCTCGCCC GGCCCCGCGGCCGACGGCCCCATCGACCTGAGCAAGAAGCCGCGGCGCCCGCTCCCCGGAGCCCCGGCACCGGCGCTGGCCGACTACCAC GAGTGCACGGCCTGCCGCGTGAGCTTCCACAGCCTCGAGGCCTACCTGGCGCACAAGAAGTACTCGTGCCCCGCTGCGCCACCGCCCGGC GCGCTCGGCCTGCCCGCCGCCGCCTGCCCCTACTGCCCCCCGAACGGCCCGGTGCGCGGGGACCTGCTGGAGCATTTCCGCCTGGCGCAC GGCCTGCTGCTCGGCGCGCCCCTGGCCGGCCCGGGGGTCGAGGCCCGGACGCCGGCCGACCGCGGCCCCTCGCCCGCTCCCGCCCCCGCC GCCTCCCCGCAGCCCGGGTCCCGTGGCCCCCGGGACGGCCTCGGCCCGGAGCCCCAGGAGCCGCCGCCCGGCCCGCCCCCGTCCCCGGCC GCCGCGCCCGAGGCCGTGCCGCCCCCGCCGGCGCCCCCCTCCTACTCGGACAAGGGCGTCCAGACTCCCAGCAAGGGCACGCCGGCGCCG CTGCCCAACGGCAACCACCGGTACTGCCGTCTTTGCAACATCAAGTTCAGCAGCCTGTCCACCTTCATCGCCCACAAGAAGTATTACTGC TCCTCGCACGCCGCCGAGCACGTGAAGTGAGCGCCCACACTACAGCCGCAGACGCTTTGCACGCCCCGCTGCGATGCGGGGAGGGGGCCG CCCCCAGGCCGCACGGACTGCCGCTCCTGGGAACCCCGCCACGCACAGGCCTCGGCGGAGGGGGCCGCAGGGGGCAGCGCCCGCCTGGAC CCTTGGCACTTAATAAAGAAGTTCAGTTTGATGAGCATGGTGGTGGGCCAGCCTAGTTCTCTGAGCCAGCAGGCACACGCAGCCAGTGTC ACATCCAGCCCTGCTGTGTACACAGGCCACTGCGGCCCGGAGTGGCTGGGCCCCTGCCGGAGTTACACCACTGGGTGCTAAGAGCTAGAC CGAGCGTCAGGTCGGAGGCCACAGCACACACTGAACCACTGCAGGGCCAGCCGCTCTGGGCCTACCCCACCCCAGCCCTGTCCATACCCC CCTAGGGAGAGCAGCCTCCACACTGCTGACCCTCTCCACGGTCTACTTGGCCGCCAGACACGGAAGAGAGGGGCACTGACCTGGCTGGCC ATCCCCCAACCCTGCATGGTCCTGGGGGTCCGATGTCCCCCATCCAGTCACAGACCACCAGGGATGGCAGGGGTGGGGGCGCCCTGGGGA GGATGAAGCCCTGTGGTGGCCAAGGCTGGGGGAAGAACAGGCGAGTAGGTGGACAGGAGAGGGAGACCGAGGCCGAAGCATTCCCTGGCC TCGAGGGGGTCCTGATGCTCACTGGTGGGTGCATCTCTCCCTCAGAGCCAGGGCTGAGGACAACCGTGTGGTGGAGTCACCTGTGGTGGC TGAGTCGGTGCAAGGGTCAGGGCACCCCCACCCCCCGTCTTCCCTGCCCTGCTGGGCTGTATGGGCCGGCCACACTGAGGGAGTACTTGG CTGACCGCGTTCAGCCACGGTGTCACCGTGCCGTTTGCAGCTGGGTGGCTGTGGGCAGCCTCCAGCGTGCTTTCTCACTGGAGAACAAAA ACACTCTTCTATTCCCAACGTCACTTTATTTTTTTATTTTTGGGTCTCACTCTCTTGCCCAGGCTGGAGTCCCTGGGATACTCACGGCTC ATTGCAGCCTTGACCTCGTGAGCTCAAGTGATCTTCCCACCTCAGTTTCCTCAGTAGCTGGGGCTACAGGTGTACACCACCATGCCCAGC TAGTTTTTGTGTTTTTTGTAGAAATGGTGTCTCCCTATGTTGCCCAGGCTGGGCAACCCTATATTTTTTAGAGCTAAGGTCTTGTTCGGT TCCCCAGGCTGAAGTTCAGTGGCACGATCATAAGTCACTGCAGCCTCAAACTCCTGAGCTGAAACCATCTTCCTGCCTCAGCCTCCCAAG TAGCTGGGACTACAGGCTTGACCCACCACACCTGGCTAACTTTTAAAACATTTTTATAGAGACAGGGTCTTCCTGTGTTGCCCAAGATGG TCTCAAACTCCTGACCTCCAGTGCTCCTGCAGCCTCAGCCTCCCAGAACATTAGGATGACAGGCGCGAGCTACCATTCCCAGCCCATTTC CAACTCTACTTGAACTAGCTGCATTTTTTTCAGCTTTGCCCAGCAGCCCTGGTGAGTGCAGGCAAAGAAGGCAGCGGCCTCTTGCCTCAG AAGCCCGAGGCACTTCCCAGCCAGTCCAGGGCTCGCTCTCCTGCCCGCAGCTCGTGGGCCTGGGTCTGCAGCGCTTTCAGTGGTCTCTGA TGTCTTTGTCCACCTGCCCACTAGGGTCTGGCCAGATGGGGCCCGTAGCCAAGGCCAATGGACAGACATGGCTTCCACCCCCTCCCAGGG ACCTGAGCTCCAGGGAAGGCAAGCAGCCTTTTCCTCACAGACACCCCTTCTGTTGAAGACCAGGTGATAGCGGGGACGGGAGGCACGGAC CCCTCAAGTCTGCTCATCATTTGCATTGTTGTGCATGAATTTGCGTGGAGGGTTCAGGAGGCTCTGCCCCCAAGTCCTCCCCAGGTGCGG GCGCAGGGTACAAGATGTTCTAAAAAATAGAAACCTGGTGGGGGGTTGTCACTGTGTGTACATTTAAAAGTAAACTGTTTAAAGGTG >16843_16843_2_CIRH1A-ZFPM1_CIRH1A_chr16_69201088_ENST00000352319_ZFPM1_chr16_88593221_ENST00000319555_length(amino acids)=1405AA_BP=1035 MGEFKVHRVRFFNYVPSGIRCVAYNNQSNRLAVSRTDGTVEIYNLSANYFQEKFFPGHESRATEALCWAEGQRLFSAGLNGEIMEYDLQA LNIKYAMDAFGGPIWSMAASPSGSQLLVGCEDGSVKLFQITPDKIQFERNFDRQKSRILSLSWHPSGTHIAAGSIDYISVFDVKSGSAVH KMIVDRQYMGVSKRKCIVWGVAFLSDGTIISVDSAGKVQFWDSATGTLVKSHLIANADVQSIAVADQEDSFVVGTAEGTVFHFQLVPVTS NSSEKQWVRTKPFQHHTHDVRTVAHSPTALISGGTDTHLVFRPLMEKVEVKNYDAALRKITFPHRCLISCSKKRQLLLFQFAHHLELWRL GSTVATGTVEAMCLLAVSPDGNWLAASGTSAGVHVYNVKQLKLHCTVPAYNFPVTAMAIAPNTNNLVIAHSDQQVFEYSIPDKQYTDWSR TVQKQGFHHLWLQRDTPITHISFHPKRPMHILLHDAYMFCIIDKSLPLPNDKTLLYNPFPPTNESDVIRRRTAHAFKISKIYKSPALTLL LVDEACWLRTLPQALTEAEANTEIHRKDDALWCRVTKPVPAGGLLSVLLTAEPHSTPGHPVKKEPAEPTCPAPAHDLQLLPQQAGMASIL ATAVINKDVFPCKDCGIWYRSERNLQAHLLYYCASRQGTGSPAAAATDEKPKETYPNERVCPFPQCRKSCPSASSLEIHMRSHSGERPFV CLICLSAFTTKANCERHLKVHTDTLSGVCHSCGFISTTRDILYSHLVTNHMVCQPGSKGEIYSPGAGHPATKLPPDSLGSFQQQHTALQG PLASADLGLAPTPSPGLDRKALAEATNGEARAEPLAQNGGSSEPPAAPRSIKVEAVEEPEAAPILGPGEPGPQAPSRTPSPRSPAPARVK AELSSPTPGSSPVPGELGLAGALFLPQYVFGPDAAPPASEILAKMSELVHSRLQQGAGAGAGGAQTGLFPGAPKGATCFECEITFSNVNN YYVHKRLYCSGRRAPEDAPAARRPKAPPGPARAPPGQPAEPDAPRSSPGPGAREEGAGGAATPEDGAGGRGSEGSQSPGSSVDDAEDDPS RTLCEACNIRFSRHETYTVHKRYYCASRHDPPPRRPAAPPGPPGPAAPPAPSPAAPVRTRRRRKLYELHAAGAPPPPPPGHAPAPESPRP GSGSGSGPGLAPARSPGPAADGPIDLSKKPRRPLPGAPAPALADYHECTACRVSFHSLEAYLAHKKYSCPAAPPPGALGLPAAACPYCPP NGPVRGDLLEHFRLAHGLLLGAPLAGPGVEARTPADRGPSPAPAPAASPQPGSRGPRDGLGPEPQEPPPGPPPSPAAAPEAVPPPPAPPS YSDKGVQTPSKGTPAPLPNGNHRYCRLCNIKFSSLSTFIAHKKYYCSSHAAEHVK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CIRH1A-ZFPM1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 794_800 | 134.0 | 1007.0 | CTBP2 | |
| Tgene | ZFPM1 | chr16:69201088 | chr16:88593221 | ENST00000319555 | 3 | 10 | 330_341 | 134.0 | 1007.0 | TACC3 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CIRH1A-ZFPM1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CIRH1A-ZFPM1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
