
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:FRMD5-ALDH1A3 (FusionGDB2 ID:HG84978TG220) |
Fusion Gene Summary for FRMD5-ALDH1A3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: FRMD5-ALDH1A3 | Fusion gene ID: hg84978tg220 | Hgene | Tgene | Gene symbol | FRMD5 | ALDH1A3 | Gene ID | 84978 | 220 |
| Gene name | FERM domain containing 5 | aldehyde dehydrogenase 1 family member A3 | |
| Synonyms | - | ALDH1A6|ALDH6|MCOP8|RALDH3 | |
| Cytomap | ('FRMD5')('ALDH1A3') 15q15.3 | 15q26.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | FERM domain-containing protein 5 | aldehyde dehydrogenase family 1 member A3acetaldehyde dehydrogenase 6aldehyde dehydrogenase 6retinaldehyde dehydrogenase 3 | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q7Z6J6 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000402883, ENST00000417257, ENST00000484674, | ||
| Fusion gene scores | * DoF score | 12 X 11 X 5=660 | 3 X 3 X 3=27 |
| # samples | 14 | 3 | |
| ** MAII score | log2(14/660*10)=-2.23703919730085 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: FRMD5 [Title/Abstract] AND ALDH1A3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | FRMD5(44487151)-ALDH1A3(101425472), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | FRMD5-ALDH1A3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. FRMD5-ALDH1A3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ALDH1A3 | GO:0002138 | retinoic acid biosynthetic process | 27759097 |
| Tgene | ALDH1A3 | GO:0042573 | retinoic acid metabolic process | 11585737 |
| Tgene | ALDH1A3 | GO:0042574 | retinal metabolic process | 11585737 |
| Tgene | ALDH1A3 | GO:0051289 | protein homotetramerization | 27759097 |
 Fusion gene breakpoints across FRMD5 (5'-gene) Fusion gene breakpoints across FRMD5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
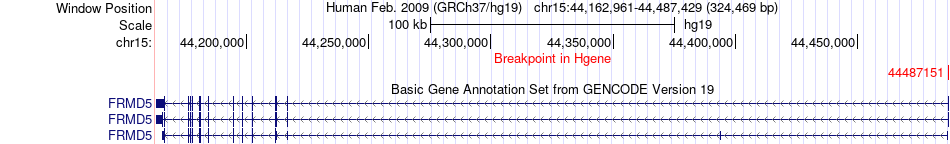 |
 Fusion gene breakpoints across ALDH1A3 (3'-gene) Fusion gene breakpoints across ALDH1A3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
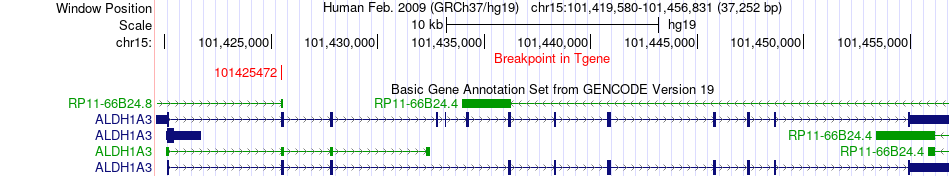 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-A486-01A | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
Top |
Fusion Gene ORF analysis for FRMD5-ALDH1A3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000402883 | ENST00000560555 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| 5CDS-3UTR | ENST00000417257 | ENST00000560555 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| 5CDS-intron | ENST00000402883 | ENST00000557963 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| 5CDS-intron | ENST00000417257 | ENST00000557963 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| In-frame | ENST00000402883 | ENST00000329841 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| In-frame | ENST00000402883 | ENST00000346623 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| In-frame | ENST00000417257 | ENST00000329841 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| In-frame | ENST00000417257 | ENST00000346623 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| intron-3CDS | ENST00000484674 | ENST00000329841 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| intron-3CDS | ENST00000484674 | ENST00000346623 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| intron-3UTR | ENST00000484674 | ENST00000560555 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
| intron-intron | ENST00000484674 | ENST00000557963 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000417257 | FRMD5 | chr15 | 44487151 | - | ENST00000329841 | ALDH1A3 | chr15 | 101425472 | + | 3572 | 279 | 87 | 1718 | 543 |
| ENST00000417257 | FRMD5 | chr15 | 44487151 | - | ENST00000346623 | ALDH1A3 | chr15 | 101425472 | + | 3251 | 279 | 87 | 1397 | 436 |
| ENST00000402883 | FRMD5 | chr15 | 44487151 | - | ENST00000329841 | ALDH1A3 | chr15 | 101425472 | + | 3551 | 258 | 66 | 1697 | 543 |
| ENST00000402883 | FRMD5 | chr15 | 44487151 | - | ENST00000346623 | ALDH1A3 | chr15 | 101425472 | + | 3230 | 258 | 66 | 1376 | 436 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000417257 | ENST00000329841 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + | 0.000396457 | 0.99960357 |
| ENST00000417257 | ENST00000346623 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + | 0.000237326 | 0.9997627 |
| ENST00000402883 | ENST00000329841 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + | 0.000395317 | 0.99960476 |
| ENST00000402883 | ENST00000346623 | FRMD5 | chr15 | 44487151 | - | ALDH1A3 | chr15 | 101425472 | + | 0.000236628 | 0.9997634 |
Top |
Fusion Genomic Features for FRMD5-ALDH1A3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| FRMD5 | chr15 | 44487150 | - | ALDH1A3 | chr15 | 101425471 | + | 0.001410391 | 0.9985896 |
| FRMD5 | chr15 | 44487150 | - | ALDH1A3 | chr15 | 101425471 | + | 0.001410391 | 0.9985896 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
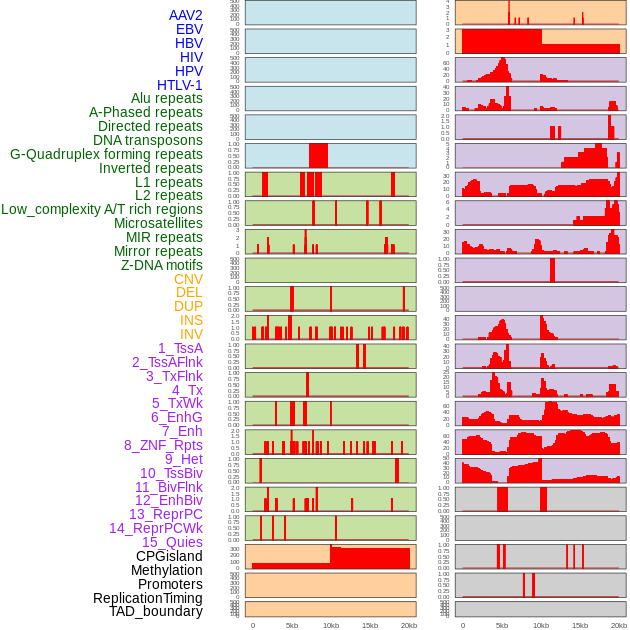 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
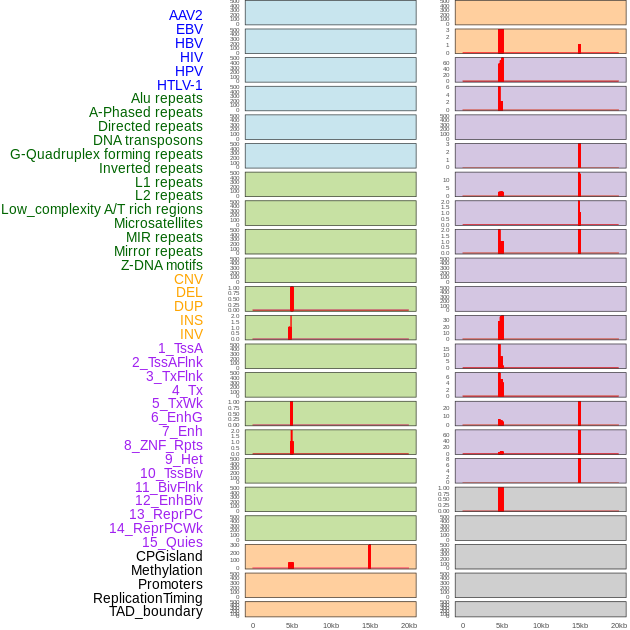 |
Top |
Fusion Protein Features for FRMD5-ALDH1A3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:44487151/chr15:101425472) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FRMD5 | . |
| FUNCTION: May be involved in regulation of cell migration (PubMed:22846708, PubMed:25448675). May regulate cell-matrix interactions via its interaction with ITGB5 and modifying ITGB5 cytoplasmic tail interactions such as with FERMT2 and TLN1. May regulate ROCK1 kinase activity possibly involved in regulation of actin stress fiber formation (PubMed:25448675). | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ALDH1A3 | chr15:44487151 | chr15:101425472 | ENST00000329841 | 0 | 13 | 257_262 | 33 | 513.0 | Nucleotide binding | NAD |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | FRMD5 | chr15:44487151 | chr15:101425472 | ENST00000417257 | - | 1 | 14 | 17_298 | 34 | 571.0 | Domain | FERM |
| Hgene | FRMD5 | chr15:44487151 | chr15:101425472 | ENST00000417257 | - | 1 | 14 | 504_524 | 34 | 571.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for FRMD5-ALDH1A3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >31352_31352_1_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000402883_ALDH1A3_chr15_101425472_ENST00000329841_length(transcript)=3551nt_BP=258nt AAGGAGGAGGAACGAGGCCAGCAGGAGGCAACGGCAGCGACGGGGCCGGGGTGATGGTGCAGGTGCCTGGGGTCGGTGCGGAGCTGCCGG GCTGAGGGACGCCTGGTCCAGGGTCCGCAGCGCCGCCGCGTCGCTCCCGGGCGGGCGGGCGGGAAGATGCTGAGCAGGTTGATGAGCGGC AGCAGCAGGAGCCTGGAGCGCGAGTACAGCTGCACCGTGCGGCTGCTGGACGACAGCGAGTACACCTGCACCATCCAGATATTTATCAAC AATGAATGGCACGAATCCAAGAGTGGGAAAAAGTTTGCTACATGTAACCCTTCAACTCGGGAGCAAATATGTGAAGTGGAAGAAGGAGAT AAGCCCGACGTGGACAAGGCTGTGGAGGCTGCACAGGTTGCCTTCCAGAGGGGCTCGCCATGGCGCCGGCTGGATGCCCTGAGTCGTGGG CGGCTGCTGCACCAGCTGGCTGACCTGGTGGAGAGGGACCGCGCCACCTTGGCCGCCCTGGAGACGATGGATACAGGGAAGCCATTTCTT CATGCTTTTTTCATCGACCTGGAGGGCTGTATTAGAACCCTCAGATACTTTGCAGGGTGGGCAGACAAAATCCAGGGCAAGACCATCCCC ACAGATGACAACGTCGTGTGCTTCACCAGGCATGAGCCCATTGGTGTCTGTGGGGCCATCACTCCATGGAACTTCCCCCTGCTGATGCTG GTGTGGAAGCTGGCACCCGCCCTCTGCTGTGGGAACACCATGGTCCTGAAGCCTGCGGAGCAGACACCTCTCACCGCCCTTTATCTCGGC TCTCTGATCAAAGAGGCCGGGTTCCCTCCAGGAGTGGTGAACATTGTGCCAGGATTCGGGCCCACAGTGGGAGCAGCAATTTCTTCTCAC CCTCAGATCAACAAGATCGCCTTCACCGGCTCCACAGAGGTTGGAAAACTGGTTAAAGAAGCTGCGTCCCGGAGCAATCTGAAGCGGGTG ACGCTGGAGCTGGGGGGGAAGAACCCCTGCATCGTGTGTGCGGACGCTGACTTGGACTTGGCAGTGGAGTGTGCCCATCAGGGAGTGTTC TTCAACCAAGGCCAGTGTTGCACGGCAGCCTCCAGGGTGTTCGTGGAGGAGCAGGTCTACTCTGAGTTTGTCAGGCGGAGCGTGGAGTAT GCCAAGAAACGGCCCGTGGGAGACCCCTTCGATGTCAAAACAGAACAGGGGCCTCAGATTGATCAAAAGCAGTTCGACAAAATCTTAGAG CTGATCGAGAGTGGGAAGAAGGAAGGGGCCAAGCTGGAATGCGGGGGCTCAGCCATGGAAGACAAGGGGCTCTTCATCAAACCCACTGTC TTCTCAGAAGTCACAGACAACATGCGGATTGCCAAAGAGGAGATTTTCGGGCCAGTGCAACCAATACTGAAGTTCAAAAGTATCGAAGAA GTGATAAAAAGAGCGAATAGCACCGACTATGGACTCACAGCAGCCGTGTTCACAAAAAATCTCGACAAAGCCCTGAAGTTGGCTTCTGCC TTAGAGTCTGGAACGGTCTGGATCAACTGCTACAACGCCCTCTATGCACAGGCTCCATTTGGTGGCTTTAAAATGTCAGGAAATGGCAGA GAACTAGGTGAATACGCTTTGGCCGAATACACAGAAGTGAAAACTGTCACCATCAAACTTGGCGACAAGAACCCCTGAAGGAAAGGCGGG GCTCCTTCCTCAAACATCGGACGGCGGAATGTGGCAGATGAAATGTGCTGGAGGAAAAAAATGACATTTCTGACCTTCCCGGGACACATT CTTCTGGAGGCTTTACATCTACTGGAGTTGAATGATTGCTGTTTTCCTCTCACTCTCCTGTTTATTCACCAGACTGGGGATGCCTATAGG TTGTCTGTGAAATCGCAGTCCTGCCTGGGGAGGGAGCTGTTGGCCATTTCTGTGTTTCCCTTTAAACCAGATCCTGGAGACAGTGAGATA CTCAGGGCGTTGTTAACAGGGAGTGGTATTTGAAGTGTCCAGCAGTTGCTTGAAATGCTTTGCCGAATCTGACTCCAGTAAGAATGTGGG AAAACCCCCTGTGTGTTCTGCAAGCAGGGCTCTTGCACCAGCGGTCTCCTCAGGGTGGACCTGCTTACAGAGCAAGCCACGCCTCTTTCC GAGGTGAAGGTGGGACCATTCCTTGGGAAAGGATTCACAGTAAGGTTTTTTGGTTTTTGTTTTTTGTTTTCTTGTTTTTAAAAAAAGGAT TTCACAGTGAGAAAGTTTTGGTTAGTGCATACCGTGGAAGGGCGCCAGGGTCTTTGTGGATTGCATGTTGACATTGACCGTGAGATTCGG CTTCAAACCAATACTGCCTTTGGAATATGACAGAATCAATAGCCCAGAGAGCTTAGTCAAAGACGATATCACGGTCTACCTTAACCAAGG CACTTTCTTAAGCAGAAAATATTGTTGAGGTTACCTTTGCTGCTAAAGATCCAATCTTCTAACGCCACAACAGCATAGCAAATCCTAGGA TAATTCACCTCCTCATTTGACAAATCAGAGCTGTAATTCGCTTTAACAAATTACGCATTTCTATCACGTTCACTAACAGCTTATGATAAG TCTGTGTAGTCTTCCTTTTCTCCAGTTCTGTTACCCAATTTAGATTAGTAAAGCGTACACAACTGGAAAGACTGCTGTAATAACACAGCC TTGTTATTTTTAAGTCCTATTTTGATATTAATTTCTGATTAGTTAGTAAATAACACCTGGATTCTATGGAGGACCTCGGTCTTCATCCAA GTGGCCTGAGTATTTCACTGGCAGGTTGTGAATTTTTCTTTTCCTCTTTGGGGATCCAAATGATGATGTGCAATTTCATGTTTTAACTTG GGAAACTGAAAGTGTTCCCATATAGCTTCAAAAACAAAAACAAATGTGTTATCCGACGGATACTTTTATGGTTACTAACTAGTACTTTCC TAATTGGGAAAGTAGTGCTTAAGTTTGCAAATTAAGTTGGGGAGGGCAATAATAAAATGAGGGCCCGTAACAGAACCAGTGTGTGTATAA CGAAAACCATGTATAAAATGGGCCTATCACCCTTGTCAGAGATATAAATTACCACATTTGCCTTCCCTTCATCAGCTAACACTTATCACT TATACTACCAATAACTTGTTAAATCAGGATTTGGCTTCATACACTGAATTTTCAGTATTTTATCTCAAGTAGATATAGACACTAACCTTG ATAGTGATACGTTAGAGGGTTCCTATTCTTCCATTGTACGATAATGTCTTTAATATGAAATGCTACATTATTTATAATTGGTAGAGTTAT TGTATCTTTTTATAGTTGTAAGTACACAGAGGTGGTATATTTAAACTTCTGTAATATACTGTATTTAGAAATGGAAATATATATAGTGTT AGGTTTCACTTCTTTTAAGGTTTACCCCTGTGGTGTGGTTTAAAAATCTATAGGCCTGGGAATTCCGATCCTAGCTGCAGATCGCATCCC ACAATGCGAGAATGATAAAATAAAATTGGATATTTGAGAAA >31352_31352_1_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000402883_ALDH1A3_chr15_101425472_ENST00000329841_length(amino acids)=543AA_BP=64 MGSVRSCRAEGRLVQGPQRRRVAPGRAGGKMLSRLMSGSSRSLEREYSCTVRLLDDSEYTCTIQIFINNEWHESKSGKKFATCNPSTREQ ICEVEEGDKPDVDKAVEAAQVAFQRGSPWRRLDALSRGRLLHQLADLVERDRATLAALETMDTGKPFLHAFFIDLEGCIRTLRYFAGWAD KIQGKTIPTDDNVVCFTRHEPIGVCGAITPWNFPLLMLVWKLAPALCCGNTMVLKPAEQTPLTALYLGSLIKEAGFPPGVVNIVPGFGPT VGAAISSHPQINKIAFTGSTEVGKLVKEAASRSNLKRVTLELGGKNPCIVCADADLDLAVECAHQGVFFNQGQCCTAASRVFVEEQVYSE FVRRSVEYAKKRPVGDPFDVKTEQGPQIDQKQFDKILELIESGKKEGAKLECGGSAMEDKGLFIKPTVFSEVTDNMRIAKEEIFGPVQPI LKFKSIEEVIKRANSTDYGLTAAVFTKNLDKALKLASALESGTVWINCYNALYAQAPFGGFKMSGNGRELGEYALAEYTEVKTVTIKLGD KNP -------------------------------------------------------------- >31352_31352_2_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000402883_ALDH1A3_chr15_101425472_ENST00000346623_length(transcript)=3230nt_BP=258nt AAGGAGGAGGAACGAGGCCAGCAGGAGGCAACGGCAGCGACGGGGCCGGGGTGATGGTGCAGGTGCCTGGGGTCGGTGCGGAGCTGCCGG GCTGAGGGACGCCTGGTCCAGGGTCCGCAGCGCCGCCGCGTCGCTCCCGGGCGGGCGGGCGGGAAGATGCTGAGCAGGTTGATGAGCGGC AGCAGCAGGAGCCTGGAGCGCGAGTACAGCTGCACCGTGCGGCTGCTGGACGACAGCGAGTACACCTGCACCATCCAGATATTTATCAAC AATGAATGGCACGAATCCAAGAGTGGGAAAAAGTTTGCTACATGTAACCCTTCAACTCGGGAGCAAATATGTGAAGTGGAAGAAGGAGAT AAGCCCGACGTGGACAAGGCTGTGGAGGCTGCACAGGTTGCCTTCCAGAGGGGCTCGCCATGGCGCCGGCTGGATGCCCTGAGTCGTGGG CGGCTGCTGCACCAGCTGGCTGACCTGGTGGAGAGGGACCGCGCCACCTTGGCCGCCGGGTTCCCTCCAGGAGTGGTGAACATTGTGCCA GGATTCGGGCCCACAGTGGGAGCAGCAATTTCTTCTCACCCTCAGATCAACAAGATCGCCTTCACCGGCTCCACAGAGGTTGGAAAACTG GTTAAAGAAGCTGCGTCCCGGAGCAATCTGAAGCGGGTGACGCTGGAGCTGGGGGGGAAGAACCCCTGCATCGTGTGTGCGGACGCTGAC TTGGACTTGGCAGTGGAGTGTGCCCATCAGGGAGTGTTCTTCAACCAAGGCCAGTGTTGCACGGCAGCCTCCAGGGTGTTCGTGGAGGAG CAGGTCTACTCTGAGTTTGTCAGGCGGAGCGTGGAGTATGCCAAGAAACGGCCCGTGGGAGACCCCTTCGATGTCAAAACAGAACAGGGG CCTCAGATTGATCAAAAGCAGTTCGACAAAATCTTAGAGCTGATCGAGAGTGGGAAGAAGGAAGGGGCCAAGCTGGAATGCGGGGGCTCA GCCATGGAAGACAAGGGGCTCTTCATCAAACCCACTGTCTTCTCAGAAGTCACAGACAACATGCGGATTGCCAAAGAGGAGATTTTCGGG CCAGTGCAACCAATACTGAAGTTCAAAAGTATCGAAGAAGTGATAAAAAGAGCGAATAGCACCGACTATGGACTCACAGCAGCCGTGTTC ACAAAAAATCTCGACAAAGCCCTGAAGTTGGCTTCTGCCTTAGAGTCTGGAACGGTCTGGATCAACTGCTACAACGCCCTCTATGCACAG GCTCCATTTGGTGGCTTTAAAATGTCAGGAAATGGCAGAGAACTAGGTGAATACGCTTTGGCCGAATACACAGAAGTGAAAACTGTCACC ATCAAACTTGGCGACAAGAACCCCTGAAGGAAAGGCGGGGCTCCTTCCTCAAACATCGGACGGCGGAATGTGGCAGATGAAATGTGCTGG AGGAAAAAAATGACATTTCTGACCTTCCCGGGACACATTCTTCTGGAGGCTTTACATCTACTGGAGTTGAATGATTGCTGTTTTCCTCTC ACTCTCCTGTTTATTCACCAGACTGGGGATGCCTATAGGTTGTCTGTGAAATCGCAGTCCTGCCTGGGGAGGGAGCTGTTGGCCATTTCT GTGTTTCCCTTTAAACCAGATCCTGGAGACAGTGAGATACTCAGGGCGTTGTTAACAGGGAGTGGTATTTGAAGTGTCCAGCAGTTGCTT GAAATGCTTTGCCGAATCTGACTCCAGTAAGAATGTGGGAAAACCCCCTGTGTGTTCTGCAAGCAGGGCTCTTGCACCAGCGGTCTCCTC AGGGTGGACCTGCTTACAGAGCAAGCCACGCCTCTTTCCGAGGTGAAGGTGGGACCATTCCTTGGGAAAGGATTCACAGTAAGGTTTTTT GGTTTTTGTTTTTTGTTTTCTTGTTTTTAAAAAAAGGATTTCACAGTGAGAAAGTTTTGGTTAGTGCATACCGTGGAAGGGCGCCAGGGT CTTTGTGGATTGCATGTTGACATTGACCGTGAGATTCGGCTTCAAACCAATACTGCCTTTGGAATATGACAGAATCAATAGCCCAGAGAG CTTAGTCAAAGACGATATCACGGTCTACCTTAACCAAGGCACTTTCTTAAGCAGAAAATATTGTTGAGGTTACCTTTGCTGCTAAAGATC CAATCTTCTAACGCCACAACAGCATAGCAAATCCTAGGATAATTCACCTCCTCATTTGACAAATCAGAGCTGTAATTCGCTTTAACAAAT TACGCATTTCTATCACGTTCACTAACAGCTTATGATAAGTCTGTGTAGTCTTCCTTTTCTCCAGTTCTGTTACCCAATTTAGATTAGTAA AGCGTACACAACTGGAAAGACTGCTGTAATAACACAGCCTTGTTATTTTTAAGTCCTATTTTGATATTAATTTCTGATTAGTTAGTAAAT AACACCTGGATTCTATGGAGGACCTCGGTCTTCATCCAAGTGGCCTGAGTATTTCACTGGCAGGTTGTGAATTTTTCTTTTCCTCTTTGG GGATCCAAATGATGATGTGCAATTTCATGTTTTAACTTGGGAAACTGAAAGTGTTCCCATATAGCTTCAAAAACAAAAACAAATGTGTTA TCCGACGGATACTTTTATGGTTACTAACTAGTACTTTCCTAATTGGGAAAGTAGTGCTTAAGTTTGCAAATTAAGTTGGGGAGGGCAATA ATAAAATGAGGGCCCGTAACAGAACCAGTGTGTGTATAACGAAAACCATGTATAAAATGGGCCTATCACCCTTGTCAGAGATATAAATTA CCACATTTGCCTTCCCTTCATCAGCTAACACTTATCACTTATACTACCAATAACTTGTTAAATCAGGATTTGGCTTCATACACTGAATTT TCAGTATTTTATCTCAAGTAGATATAGACACTAACCTTGATAGTGATACGTTAGAGGGTTCCTATTCTTCCATTGTACGATAATGTCTTT AATATGAAATGCTACATTATTTATAATTGGTAGAGTTATTGTATCTTTTTATAGTTGTAAGTACACAGAGGTGGTATATTTAAACTTCTG TAATATACTGTATTTAGAAATGGAAATATATATAGTGTTAGGTTTCACTTCTTTTAAGGTTTACCCCTGTGGTGTGGTTTAAAAATCTAT AGGCCTGGGAATTCCGATCCTAGCTGCAGATCGCATCCCACAATGCGAGAATGATAAAATAAAATTGGATATTTGAGAAA >31352_31352_2_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000402883_ALDH1A3_chr15_101425472_ENST00000346623_length(amino acids)=436AA_BP=64 MGSVRSCRAEGRLVQGPQRRRVAPGRAGGKMLSRLMSGSSRSLEREYSCTVRLLDDSEYTCTIQIFINNEWHESKSGKKFATCNPSTREQ ICEVEEGDKPDVDKAVEAAQVAFQRGSPWRRLDALSRGRLLHQLADLVERDRATLAAGFPPGVVNIVPGFGPTVGAAISSHPQINKIAFT GSTEVGKLVKEAASRSNLKRVTLELGGKNPCIVCADADLDLAVECAHQGVFFNQGQCCTAASRVFVEEQVYSEFVRRSVEYAKKRPVGDP FDVKTEQGPQIDQKQFDKILELIESGKKEGAKLECGGSAMEDKGLFIKPTVFSEVTDNMRIAKEEIFGPVQPILKFKSIEEVIKRANSTD YGLTAAVFTKNLDKALKLASALESGTVWINCYNALYAQAPFGGFKMSGNGRELGEYALAEYTEVKTVTIKLGDKNP -------------------------------------------------------------- >31352_31352_3_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000417257_ALDH1A3_chr15_101425472_ENST00000329841_length(transcript)=3572nt_BP=279nt GGCTGAGGGAAGGAGGAGGATAAGGAGGAGGAACGAGGCCAGCAGGAGGCAACGGCAGCGACGGGGCCGGGGTGATGGTGCAGGTGCCTG GGGTCGGTGCGGAGCTGCCGGGCTGAGGGACGCCTGGTCCAGGGTCCGCAGCGCCGCCGCGTCGCTCCCGGGCGGGCGGGCGGGAAGATG CTGAGCAGGTTGATGAGCGGCAGCAGCAGGAGCCTGGAGCGCGAGTACAGCTGCACCGTGCGGCTGCTGGACGACAGCGAGTACACCTGC ACCATCCAGATATTTATCAACAATGAATGGCACGAATCCAAGAGTGGGAAAAAGTTTGCTACATGTAACCCTTCAACTCGGGAGCAAATA TGTGAAGTGGAAGAAGGAGATAAGCCCGACGTGGACAAGGCTGTGGAGGCTGCACAGGTTGCCTTCCAGAGGGGCTCGCCATGGCGCCGG CTGGATGCCCTGAGTCGTGGGCGGCTGCTGCACCAGCTGGCTGACCTGGTGGAGAGGGACCGCGCCACCTTGGCCGCCCTGGAGACGATG GATACAGGGAAGCCATTTCTTCATGCTTTTTTCATCGACCTGGAGGGCTGTATTAGAACCCTCAGATACTTTGCAGGGTGGGCAGACAAA ATCCAGGGCAAGACCATCCCCACAGATGACAACGTCGTGTGCTTCACCAGGCATGAGCCCATTGGTGTCTGTGGGGCCATCACTCCATGG AACTTCCCCCTGCTGATGCTGGTGTGGAAGCTGGCACCCGCCCTCTGCTGTGGGAACACCATGGTCCTGAAGCCTGCGGAGCAGACACCT CTCACCGCCCTTTATCTCGGCTCTCTGATCAAAGAGGCCGGGTTCCCTCCAGGAGTGGTGAACATTGTGCCAGGATTCGGGCCCACAGTG GGAGCAGCAATTTCTTCTCACCCTCAGATCAACAAGATCGCCTTCACCGGCTCCACAGAGGTTGGAAAACTGGTTAAAGAAGCTGCGTCC CGGAGCAATCTGAAGCGGGTGACGCTGGAGCTGGGGGGGAAGAACCCCTGCATCGTGTGTGCGGACGCTGACTTGGACTTGGCAGTGGAG TGTGCCCATCAGGGAGTGTTCTTCAACCAAGGCCAGTGTTGCACGGCAGCCTCCAGGGTGTTCGTGGAGGAGCAGGTCTACTCTGAGTTT GTCAGGCGGAGCGTGGAGTATGCCAAGAAACGGCCCGTGGGAGACCCCTTCGATGTCAAAACAGAACAGGGGCCTCAGATTGATCAAAAG CAGTTCGACAAAATCTTAGAGCTGATCGAGAGTGGGAAGAAGGAAGGGGCCAAGCTGGAATGCGGGGGCTCAGCCATGGAAGACAAGGGG CTCTTCATCAAACCCACTGTCTTCTCAGAAGTCACAGACAACATGCGGATTGCCAAAGAGGAGATTTTCGGGCCAGTGCAACCAATACTG AAGTTCAAAAGTATCGAAGAAGTGATAAAAAGAGCGAATAGCACCGACTATGGACTCACAGCAGCCGTGTTCACAAAAAATCTCGACAAA GCCCTGAAGTTGGCTTCTGCCTTAGAGTCTGGAACGGTCTGGATCAACTGCTACAACGCCCTCTATGCACAGGCTCCATTTGGTGGCTTT AAAATGTCAGGAAATGGCAGAGAACTAGGTGAATACGCTTTGGCCGAATACACAGAAGTGAAAACTGTCACCATCAAACTTGGCGACAAG AACCCCTGAAGGAAAGGCGGGGCTCCTTCCTCAAACATCGGACGGCGGAATGTGGCAGATGAAATGTGCTGGAGGAAAAAAATGACATTT CTGACCTTCCCGGGACACATTCTTCTGGAGGCTTTACATCTACTGGAGTTGAATGATTGCTGTTTTCCTCTCACTCTCCTGTTTATTCAC CAGACTGGGGATGCCTATAGGTTGTCTGTGAAATCGCAGTCCTGCCTGGGGAGGGAGCTGTTGGCCATTTCTGTGTTTCCCTTTAAACCA GATCCTGGAGACAGTGAGATACTCAGGGCGTTGTTAACAGGGAGTGGTATTTGAAGTGTCCAGCAGTTGCTTGAAATGCTTTGCCGAATC TGACTCCAGTAAGAATGTGGGAAAACCCCCTGTGTGTTCTGCAAGCAGGGCTCTTGCACCAGCGGTCTCCTCAGGGTGGACCTGCTTACA GAGCAAGCCACGCCTCTTTCCGAGGTGAAGGTGGGACCATTCCTTGGGAAAGGATTCACAGTAAGGTTTTTTGGTTTTTGTTTTTTGTTT TCTTGTTTTTAAAAAAAGGATTTCACAGTGAGAAAGTTTTGGTTAGTGCATACCGTGGAAGGGCGCCAGGGTCTTTGTGGATTGCATGTT GACATTGACCGTGAGATTCGGCTTCAAACCAATACTGCCTTTGGAATATGACAGAATCAATAGCCCAGAGAGCTTAGTCAAAGACGATAT CACGGTCTACCTTAACCAAGGCACTTTCTTAAGCAGAAAATATTGTTGAGGTTACCTTTGCTGCTAAAGATCCAATCTTCTAACGCCACA ACAGCATAGCAAATCCTAGGATAATTCACCTCCTCATTTGACAAATCAGAGCTGTAATTCGCTTTAACAAATTACGCATTTCTATCACGT TCACTAACAGCTTATGATAAGTCTGTGTAGTCTTCCTTTTCTCCAGTTCTGTTACCCAATTTAGATTAGTAAAGCGTACACAACTGGAAA GACTGCTGTAATAACACAGCCTTGTTATTTTTAAGTCCTATTTTGATATTAATTTCTGATTAGTTAGTAAATAACACCTGGATTCTATGG AGGACCTCGGTCTTCATCCAAGTGGCCTGAGTATTTCACTGGCAGGTTGTGAATTTTTCTTTTCCTCTTTGGGGATCCAAATGATGATGT GCAATTTCATGTTTTAACTTGGGAAACTGAAAGTGTTCCCATATAGCTTCAAAAACAAAAACAAATGTGTTATCCGACGGATACTTTTAT GGTTACTAACTAGTACTTTCCTAATTGGGAAAGTAGTGCTTAAGTTTGCAAATTAAGTTGGGGAGGGCAATAATAAAATGAGGGCCCGTA ACAGAACCAGTGTGTGTATAACGAAAACCATGTATAAAATGGGCCTATCACCCTTGTCAGAGATATAAATTACCACATTTGCCTTCCCTT CATCAGCTAACACTTATCACTTATACTACCAATAACTTGTTAAATCAGGATTTGGCTTCATACACTGAATTTTCAGTATTTTATCTCAAG TAGATATAGACACTAACCTTGATAGTGATACGTTAGAGGGTTCCTATTCTTCCATTGTACGATAATGTCTTTAATATGAAATGCTACATT ATTTATAATTGGTAGAGTTATTGTATCTTTTTATAGTTGTAAGTACACAGAGGTGGTATATTTAAACTTCTGTAATATACTGTATTTAGA AATGGAAATATATATAGTGTTAGGTTTCACTTCTTTTAAGGTTTACCCCTGTGGTGTGGTTTAAAAATCTATAGGCCTGGGAATTCCGAT CCTAGCTGCAGATCGCATCCCACAATGCGAGAATGATAAAATAAAATTGGATATTTGAGAAA >31352_31352_3_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000417257_ALDH1A3_chr15_101425472_ENST00000329841_length(amino acids)=543AA_BP=64 MGSVRSCRAEGRLVQGPQRRRVAPGRAGGKMLSRLMSGSSRSLEREYSCTVRLLDDSEYTCTIQIFINNEWHESKSGKKFATCNPSTREQ ICEVEEGDKPDVDKAVEAAQVAFQRGSPWRRLDALSRGRLLHQLADLVERDRATLAALETMDTGKPFLHAFFIDLEGCIRTLRYFAGWAD KIQGKTIPTDDNVVCFTRHEPIGVCGAITPWNFPLLMLVWKLAPALCCGNTMVLKPAEQTPLTALYLGSLIKEAGFPPGVVNIVPGFGPT VGAAISSHPQINKIAFTGSTEVGKLVKEAASRSNLKRVTLELGGKNPCIVCADADLDLAVECAHQGVFFNQGQCCTAASRVFVEEQVYSE FVRRSVEYAKKRPVGDPFDVKTEQGPQIDQKQFDKILELIESGKKEGAKLECGGSAMEDKGLFIKPTVFSEVTDNMRIAKEEIFGPVQPI LKFKSIEEVIKRANSTDYGLTAAVFTKNLDKALKLASALESGTVWINCYNALYAQAPFGGFKMSGNGRELGEYALAEYTEVKTVTIKLGD KNP -------------------------------------------------------------- >31352_31352_4_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000417257_ALDH1A3_chr15_101425472_ENST00000346623_length(transcript)=3251nt_BP=279nt GGCTGAGGGAAGGAGGAGGATAAGGAGGAGGAACGAGGCCAGCAGGAGGCAACGGCAGCGACGGGGCCGGGGTGATGGTGCAGGTGCCTG GGGTCGGTGCGGAGCTGCCGGGCTGAGGGACGCCTGGTCCAGGGTCCGCAGCGCCGCCGCGTCGCTCCCGGGCGGGCGGGCGGGAAGATG CTGAGCAGGTTGATGAGCGGCAGCAGCAGGAGCCTGGAGCGCGAGTACAGCTGCACCGTGCGGCTGCTGGACGACAGCGAGTACACCTGC ACCATCCAGATATTTATCAACAATGAATGGCACGAATCCAAGAGTGGGAAAAAGTTTGCTACATGTAACCCTTCAACTCGGGAGCAAATA TGTGAAGTGGAAGAAGGAGATAAGCCCGACGTGGACAAGGCTGTGGAGGCTGCACAGGTTGCCTTCCAGAGGGGCTCGCCATGGCGCCGG CTGGATGCCCTGAGTCGTGGGCGGCTGCTGCACCAGCTGGCTGACCTGGTGGAGAGGGACCGCGCCACCTTGGCCGCCGGGTTCCCTCCA GGAGTGGTGAACATTGTGCCAGGATTCGGGCCCACAGTGGGAGCAGCAATTTCTTCTCACCCTCAGATCAACAAGATCGCCTTCACCGGC TCCACAGAGGTTGGAAAACTGGTTAAAGAAGCTGCGTCCCGGAGCAATCTGAAGCGGGTGACGCTGGAGCTGGGGGGGAAGAACCCCTGC ATCGTGTGTGCGGACGCTGACTTGGACTTGGCAGTGGAGTGTGCCCATCAGGGAGTGTTCTTCAACCAAGGCCAGTGTTGCACGGCAGCC TCCAGGGTGTTCGTGGAGGAGCAGGTCTACTCTGAGTTTGTCAGGCGGAGCGTGGAGTATGCCAAGAAACGGCCCGTGGGAGACCCCTTC GATGTCAAAACAGAACAGGGGCCTCAGATTGATCAAAAGCAGTTCGACAAAATCTTAGAGCTGATCGAGAGTGGGAAGAAGGAAGGGGCC AAGCTGGAATGCGGGGGCTCAGCCATGGAAGACAAGGGGCTCTTCATCAAACCCACTGTCTTCTCAGAAGTCACAGACAACATGCGGATT GCCAAAGAGGAGATTTTCGGGCCAGTGCAACCAATACTGAAGTTCAAAAGTATCGAAGAAGTGATAAAAAGAGCGAATAGCACCGACTAT GGACTCACAGCAGCCGTGTTCACAAAAAATCTCGACAAAGCCCTGAAGTTGGCTTCTGCCTTAGAGTCTGGAACGGTCTGGATCAACTGC TACAACGCCCTCTATGCACAGGCTCCATTTGGTGGCTTTAAAATGTCAGGAAATGGCAGAGAACTAGGTGAATACGCTTTGGCCGAATAC ACAGAAGTGAAAACTGTCACCATCAAACTTGGCGACAAGAACCCCTGAAGGAAAGGCGGGGCTCCTTCCTCAAACATCGGACGGCGGAAT GTGGCAGATGAAATGTGCTGGAGGAAAAAAATGACATTTCTGACCTTCCCGGGACACATTCTTCTGGAGGCTTTACATCTACTGGAGTTG AATGATTGCTGTTTTCCTCTCACTCTCCTGTTTATTCACCAGACTGGGGATGCCTATAGGTTGTCTGTGAAATCGCAGTCCTGCCTGGGG AGGGAGCTGTTGGCCATTTCTGTGTTTCCCTTTAAACCAGATCCTGGAGACAGTGAGATACTCAGGGCGTTGTTAACAGGGAGTGGTATT TGAAGTGTCCAGCAGTTGCTTGAAATGCTTTGCCGAATCTGACTCCAGTAAGAATGTGGGAAAACCCCCTGTGTGTTCTGCAAGCAGGGC TCTTGCACCAGCGGTCTCCTCAGGGTGGACCTGCTTACAGAGCAAGCCACGCCTCTTTCCGAGGTGAAGGTGGGACCATTCCTTGGGAAA GGATTCACAGTAAGGTTTTTTGGTTTTTGTTTTTTGTTTTCTTGTTTTTAAAAAAAGGATTTCACAGTGAGAAAGTTTTGGTTAGTGCAT ACCGTGGAAGGGCGCCAGGGTCTTTGTGGATTGCATGTTGACATTGACCGTGAGATTCGGCTTCAAACCAATACTGCCTTTGGAATATGA CAGAATCAATAGCCCAGAGAGCTTAGTCAAAGACGATATCACGGTCTACCTTAACCAAGGCACTTTCTTAAGCAGAAAATATTGTTGAGG TTACCTTTGCTGCTAAAGATCCAATCTTCTAACGCCACAACAGCATAGCAAATCCTAGGATAATTCACCTCCTCATTTGACAAATCAGAG CTGTAATTCGCTTTAACAAATTACGCATTTCTATCACGTTCACTAACAGCTTATGATAAGTCTGTGTAGTCTTCCTTTTCTCCAGTTCTG TTACCCAATTTAGATTAGTAAAGCGTACACAACTGGAAAGACTGCTGTAATAACACAGCCTTGTTATTTTTAAGTCCTATTTTGATATTA ATTTCTGATTAGTTAGTAAATAACACCTGGATTCTATGGAGGACCTCGGTCTTCATCCAAGTGGCCTGAGTATTTCACTGGCAGGTTGTG AATTTTTCTTTTCCTCTTTGGGGATCCAAATGATGATGTGCAATTTCATGTTTTAACTTGGGAAACTGAAAGTGTTCCCATATAGCTTCA AAAACAAAAACAAATGTGTTATCCGACGGATACTTTTATGGTTACTAACTAGTACTTTCCTAATTGGGAAAGTAGTGCTTAAGTTTGCAA ATTAAGTTGGGGAGGGCAATAATAAAATGAGGGCCCGTAACAGAACCAGTGTGTGTATAACGAAAACCATGTATAAAATGGGCCTATCAC CCTTGTCAGAGATATAAATTACCACATTTGCCTTCCCTTCATCAGCTAACACTTATCACTTATACTACCAATAACTTGTTAAATCAGGAT TTGGCTTCATACACTGAATTTTCAGTATTTTATCTCAAGTAGATATAGACACTAACCTTGATAGTGATACGTTAGAGGGTTCCTATTCTT CCATTGTACGATAATGTCTTTAATATGAAATGCTACATTATTTATAATTGGTAGAGTTATTGTATCTTTTTATAGTTGTAAGTACACAGA GGTGGTATATTTAAACTTCTGTAATATACTGTATTTAGAAATGGAAATATATATAGTGTTAGGTTTCACTTCTTTTAAGGTTTACCCCTG TGGTGTGGTTTAAAAATCTATAGGCCTGGGAATTCCGATCCTAGCTGCAGATCGCATCCCACAATGCGAGAATGATAAAATAAAATTGGA TATTTGAGAAA >31352_31352_4_FRMD5-ALDH1A3_FRMD5_chr15_44487151_ENST00000417257_ALDH1A3_chr15_101425472_ENST00000346623_length(amino acids)=436AA_BP=64 MGSVRSCRAEGRLVQGPQRRRVAPGRAGGKMLSRLMSGSSRSLEREYSCTVRLLDDSEYTCTIQIFINNEWHESKSGKKFATCNPSTREQ ICEVEEGDKPDVDKAVEAAQVAFQRGSPWRRLDALSRGRLLHQLADLVERDRATLAAGFPPGVVNIVPGFGPTVGAAISSHPQINKIAFT GSTEVGKLVKEAASRSNLKRVTLELGGKNPCIVCADADLDLAVECAHQGVFFNQGQCCTAASRVFVEEQVYSEFVRRSVEYAKKRPVGDP FDVKTEQGPQIDQKQFDKILELIESGKKEGAKLECGGSAMEDKGLFIKPTVFSEVTDNMRIAKEEIFGPVQPILKFKSIEEVIKRANSTD YGLTAAVFTKNLDKALKLASALESGTVWINCYNALYAQAPFGGFKMSGNGRELGEYALAEYTEVKTVTIKLGDKNP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for FRMD5-ALDH1A3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | FRMD5 | chr15:44487151 | chr15:101425472 | ENST00000417257 | - | 1 | 14 | 308_353 | 34.0 | 571.0 | ROCK1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for FRMD5-ALDH1A3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for FRMD5-ALDH1A3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C3554524 | MICROPHTHALMIA, ISOLATED 8 | 10 | CTD_human;GENOMICS_ENGLAND;UNIPROT | |
| Tgene | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human | |
| Tgene | C0025202 | melanoma | 1 | CTD_human | |
| Tgene | C0038356 | Stomach Neoplasms | 1 | CTD_human | |
| Tgene | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human | |
| Tgene | C1855052 | MICROPHTHALMIA, ISOLATED 1 | 1 | ORPHANET | |
| Tgene | C4551977 | Microphthalmos, Autosomal Recessive | 1 | ORPHANET |
