
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:API5-LCMT1 (FusionGDB2 ID:HG8539TG51451) |
Fusion Gene Summary for API5-LCMT1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: API5-LCMT1 | Fusion gene ID: hg8539tg51451 | Hgene | Tgene | Gene symbol | API5 | LCMT1 | Gene ID | 8539 | 51451 |
| Gene name | apoptosis inhibitor 5 | leucine carboxyl methyltransferase 1 | |
| Synonyms | AAC-11|AAC11 | CGI-68|LCMT|PPMT1 | |
| Cytomap | ('API5')('LCMT1') 11p12 | 16p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | apoptosis inhibitor 5FIFantiapoptosis clone 11 proteincell migration-inducing gene 8 proteinfibroblast growth factor 2-interacting factor 2migration-inducing protein MIG8 | leucine carboxyl methyltransferase 1[Phosphatase 2A protein]-leucine-carboxy methyltransferase 1protein phosphatase methyltransferase 1protein-leucine O-methyltransferase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000529334, ENST00000378852, ENST00000420461, ENST00000455725, ENST00000531273, ENST00000534600, ENST00000534695, | ||
| Fusion gene scores | * DoF score | 7 X 5 X 5=175 | 5 X 7 X 4=140 |
| # samples | 7 | 6 | |
| ** MAII score | log2(7/175*10)=-1.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/140*10)=-1.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: API5 [Title/Abstract] AND LCMT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | API5(43357544)-LCMT1(25172422), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | API5-LCMT1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. API5-LCMT1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. API5-LCMT1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. API5-LCMT1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. API5-LCMT1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. API5-LCMT1 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. API5-LCMT1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | LCMT1 | GO:0006481 | C-terminal protein methylation | 10600115 |
 Fusion gene breakpoints across API5 (5'-gene) Fusion gene breakpoints across API5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
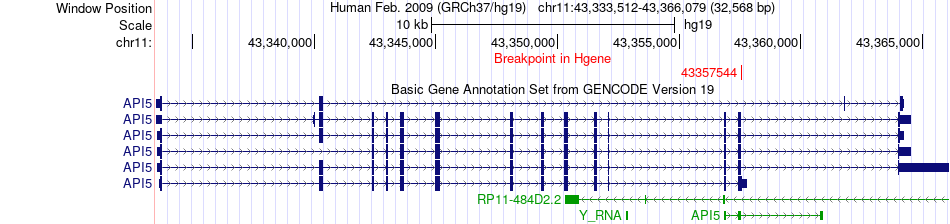 |
 Fusion gene breakpoints across LCMT1 (3'-gene) Fusion gene breakpoints across LCMT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
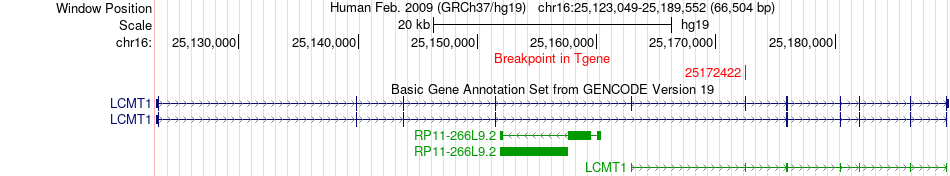 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-L5-A4OQ | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
Top |
Fusion Gene ORF analysis for API5-LCMT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000529334 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 3UTR-3UTR | ENST00000529334 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 3UTR-intron | ENST00000529334 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-3UTR | ENST00000378852 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-3UTR | ENST00000420461 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-3UTR | ENST00000455725 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-3UTR | ENST00000531273 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-3UTR | ENST00000534600 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-intron | ENST00000378852 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-intron | ENST00000420461 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-intron | ENST00000455725 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-intron | ENST00000531273 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| 5CDS-intron | ENST00000534600 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| Frame-shift | ENST00000534600 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| In-frame | ENST00000378852 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| In-frame | ENST00000420461 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| In-frame | ENST00000455725 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| In-frame | ENST00000531273 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| intron-3CDS | ENST00000534695 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| intron-3UTR | ENST00000534695 | ENST00000572869 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
| intron-intron | ENST00000534695 | ENST00000380966 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000455725 | API5 | chr11 | 43357544 | + | ENST00000399069 | LCMT1 | chr16 | 25172422 | + | 2451 | 1704 | 245 | 2242 | 665 |
| ENST00000531273 | API5 | chr11 | 43357544 | + | ENST00000399069 | LCMT1 | chr16 | 25172422 | + | 2378 | 1631 | 121 | 2169 | 682 |
| ENST00000420461 | API5 | chr11 | 43357544 | + | ENST00000399069 | LCMT1 | chr16 | 25172422 | + | 2207 | 1460 | 112 | 1998 | 628 |
| ENST00000378852 | API5 | chr11 | 43357544 | + | ENST00000399069 | LCMT1 | chr16 | 25172422 | + | 2364 | 1617 | 107 | 2155 | 682 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000455725 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.00383781 | 0.9961622 |
| ENST00000531273 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.002048279 | 0.9979518 |
| ENST00000420461 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.002047089 | 0.99795294 |
| ENST00000378852 | ENST00000399069 | API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.002078286 | 0.99792176 |
Top |
Fusion Genomic Features for API5-LCMT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.000441325 | 0.9995586 |
| API5 | chr11 | 43357544 | + | LCMT1 | chr16 | 25172422 | + | 0.000441325 | 0.9995586 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
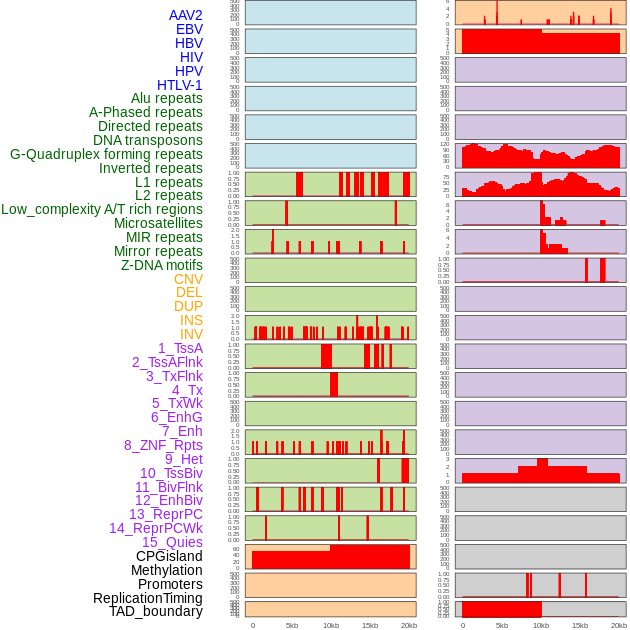 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
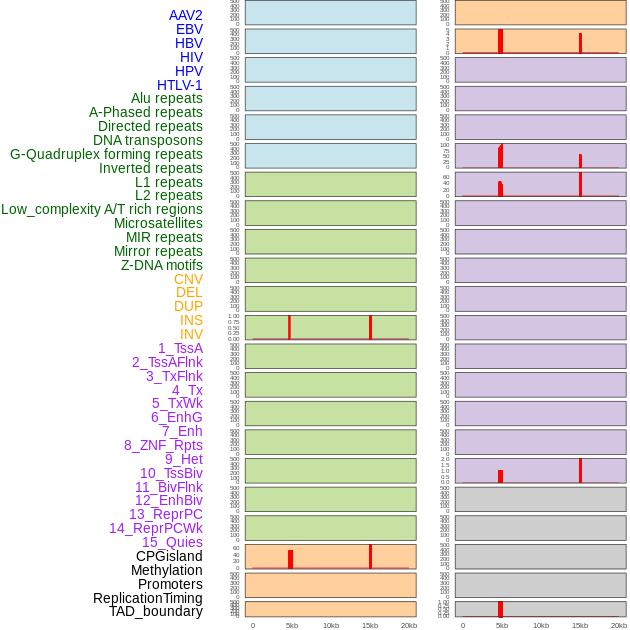 |
Top |
Fusion Protein Features for API5-LCMT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:43357544/chr16:25172422) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000378852 | + | 13 | 14 | 454_475 | 497 | 505.0 | Motif | Nuclear localization signal |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000531273 | + | 13 | 14 | 454_475 | 497 | 525.0 | Motif | Nuclear localization signal |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000378852 | + | 13 | 14 | 2_360 | 497 | 505.0 | Region | Note=ARM-like and Heat-like helical repeats |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000378852 | + | 13 | 14 | 370_391 | 497 | 505.0 | Region | Note=Leucine-zipper |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000420461 | + | 12 | 13 | 2_360 | 443 | 451.0 | Region | Note=ARM-like and Heat-like helical repeats |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000420461 | + | 12 | 13 | 370_391 | 443 | 451.0 | Region | Note=Leucine-zipper |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000531273 | + | 13 | 14 | 2_360 | 497 | 525.0 | Region | Note=ARM-like and Heat-like helical repeats |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000531273 | + | 13 | 14 | 370_391 | 497 | 525.0 | Region | Note=Leucine-zipper |
| Tgene | LCMT1 | chr11:43357544 | chr16:25172422 | ENST00000380966 | 0 | 9 | 171_172 | 0 | 280.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | LCMT1 | chr11:43357544 | chr16:25172422 | ENST00000399069 | 4 | 11 | 171_172 | 155 | 335.0 | Region | S-adenosyl-L-methionine binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | API5 | chr11:43357544 | chr16:25172422 | ENST00000420461 | + | 12 | 13 | 454_475 | 443 | 451.0 | Motif | Nuclear localization signal |
Top |
Fusion Gene Sequence for API5-LCMT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >5423_5423_1_API5-LCMT1_API5_chr11_43357544_ENST00000378852_LCMT1_chr16_25172422_ENST00000399069_length(transcript)=2364nt_BP=1617nt GCGGCTGCACTGGCGGCAGCTGGAGGTGTAATAGTGCGGGTAGTGGGTTTGGAGAAGTTCCGAGGCGGCGGTGGCGCCGGTCAGGACAAG GATAGCGGAACCGGGCCCTGGGCTTGTCGCTCACCATGCCGACAGTAGAGGAGCTTTACCGCAATTATGGCATCCTGGCCGATGCCACGG AGCAAGTGGGCCAGCATAAAGATGCCTATCAAGTGATATTGGATGGTGTGAAAGGTGGTACTAAGGAAAAGCGATTAGCAGCTCAATTTA TTCCGAAATTCTTTAAGCATTTTCCAGAATTGGCTGATTCTGCTATCAATGCACAGTTAGACCTCTGTGAGGATGAAGATGTATCTATTC GACGTCAAGCAATTAAAGAACTGCCTCAATTTGCCACTGGAGAAAATCTTCCTCGAGTGGCAGATATACTAACGCAACTTTTGCAGACAG ATGACTCTGCAGAATTTAACCTAGTGAACAATGCCCTATTAAGTATATTTAAAATGGATGCAAAAGGGACTTTAGGTGGGTTGTTCAGCC AAATACTTCAAGGAGAGGACATTGTTAGAGAACGAGCAATTAAATTCCTTTCTACAAAACTTAAGACTTTACCAGATGAAGTCTTAACAA AGGAAGTGGAAGAGCTTATACTAACTGAATCCAAAAAGGTCCTAGAAGATGTGACTGGTGAAGAATTTGTTCTATTTATGAAGATACTGT CTGGGTTAAAAAGCTTACAGACAGTGAGTGGAAGACAGCAACTTGTAGAGTTGGTGGCTGAACAGGCCGACCTAGAACAGACCTTCAATC CCTCGGATCCTGACTGTGTGGACAGGCTCTTACAGTGCACTCGGCAGGCAGTACCCCTCTTCTCTAAAAATGTCCATTCCACAAGGTTTG TGACATATTTCTGTGAGCAGGTTCTCCCTAACCTCGGTACCTTGACTACCCCAGTGGAAGGTCTTGATATACAGTTGGAGGTATTGAAAT TGTTGGCGGAGATGAGTTCATTTTGTGGTGACATGGAAAAACTAGAAACAAATTTAAGGAAACTATTTGATAAGTTATTGGAATACATGC CCCTCCCTCCAGAAGAGGCAGAAAATGGAGAGAATGCTGGTAATGAAGAACCCAAGCTACAGTTCAGTTATGTGGAATGTTTGTTGTACA GTTTTCACCAGTTGGGCCGAAAACTTCCAGATTTCTTAACAGCCAAACTGAATGCAGAAAAGCTCAAAGATTTCAAAATCAGGCTGCAGT ACTTTGCACGGGGCCTGCAAGTTTATATCAGACAACTTCGCTTAGCTCTCCAGGGTAAAACGGGTGAGGCCTTAAAAACAGAAGAGAACA AGATTAAAGTCGTTGCATTGAAAATAACAAACAATATCAATGTTTTAATCAAGGATCTCTTCCACATTCCTCCTTCTTATAAGAGCACAG TAACACTATCCTGGAAACCTGTACAAAAGGTTGAGATTGGGCAAAAGAGAGCCAGTGAAGATACAACTTCAGGTTCACCACCCAAGAAAT CTTCAGCAGGACCAAAAAGAGATGCCAGGCAGATTTATAACCCTCCCAGTGGGAAATATAGCAGCAATTTGGGCAACTTTAATTATGATG GACACATACTGGATTCAAAGAGATATGCCGTTATTGGAGCAGATCTCCGAGACCTGTCTGAACTGGAAGAGAAGCTAAAGAAATGTAACA TGAATACACAATTGCCAACACTCCTGATAGCTGAATGTGTGCTGGTTTACATGACTCCAGAGCAGTCCGCAAACCTCCTGAAGTGGGCAG CCAACAGTTTTGAGAGAGCCATGTTCATAAACTACGAACAGGTGAACATGGGTGATCGGTTTGGGCAGATCATGATTGAAAACCTGCGGA GACGCCAGTGTGACCTGGCGGGAGTGGAGACCTGCAAGTCATTAGAGTCACAGAAAGAACGGCTCCTGTCGAATGGGTGGGAAACAGCAT CGGCCGTCGACATGATGGAGTTGTACAACAGGTTACCTCGAGCTGAAGTGAGCAGGATAGAATCACTTGAATTCCTGGATGAAATGGAGC TGCTGGAGCAGCTCATGCGGCATTACTGCCTTTGCTGGGCAACCAAAGGAGGAAATGAGCTTGGGCTGAAGGAGATAACTTATTAATCTG TCGAAGGCTTATGCCGAGCCAGAAGCCGAAGCCACTTGCCCTCCTGGAGGAGACCTGCAAGCTCCCTGAGCGGTGGGCGGGCCTCGTCCG CAGGTCTCATCCCACACTCTTGAGAAGCCTTGGTCACTACAGTGGTCGCACATGTTCCTCTTCCTGTTCCTGTTGACATGTCGTTGTTTA AATAAATCTCACTTGCCACCAGTC >5423_5423_1_API5-LCMT1_API5_chr11_43357544_ENST00000378852_LCMT1_chr16_25172422_ENST00000399069_length(amino acids)=682AA_BP=503 MGLSLTMPTVEELYRNYGILADATEQVGQHKDAYQVILDGVKGGTKEKRLAAQFIPKFFKHFPELADSAINAQLDLCEDEDVSIRRQAIK ELPQFATGENLPRVADILTQLLQTDDSAEFNLVNNALLSIFKMDAKGTLGGLFSQILQGEDIVRERAIKFLSTKLKTLPDEVLTKEVEEL ILTESKKVLEDVTGEEFVLFMKILSGLKSLQTVSGRQQLVELVAEQADLEQTFNPSDPDCVDRLLQCTRQAVPLFSKNVHSTRFVTYFCE QVLPNLGTLTTPVEGLDIQLEVLKLLAEMSSFCGDMEKLETNLRKLFDKLLEYMPLPPEEAENGENAGNEEPKLQFSYVECLLYSFHQLG RKLPDFLTAKLNAEKLKDFKIRLQYFARGLQVYIRQLRLALQGKTGEALKTEENKIKVVALKITNNINVLIKDLFHIPPSYKSTVTLSWK PVQKVEIGQKRASEDTTSGSPPKKSSAGPKRDARQIYNPPSGKYSSNLGNFNYDGHILDSKRYAVIGADLRDLSELEEKLKKCNMNTQLP TLLIAECVLVYMTPEQSANLLKWAANSFERAMFINYEQVNMGDRFGQIMIENLRRRQCDLAGVETCKSLESQKERLLSNGWETASAVDMM ELYNRLPRAEVSRIESLEFLDEMELLEQLMRHYCLCWATKGGNELGLKEITY -------------------------------------------------------------- >5423_5423_2_API5-LCMT1_API5_chr11_43357544_ENST00000420461_LCMT1_chr16_25172422_ENST00000399069_length(transcript)=2207nt_BP=1460nt GACTGGCGGCTGCACTGGCGGCAGCTGGAGGTGTAATAGTGCGGGTAGTGGGTTTGGAGAAGTTCCGAGGCGGCGGTGGCGCCGGTCAGG ACAAGGATAGCGGAACCGGGCCCTGGGCTTGTCGCTCACCATGCCGACAGTAGAGGAGCTTTACCGCAATTATGGCATCCTGGCCGATGC CACGGAGCAAGTGGGCCAGATTCGACGTCAAGCAATTAAAGAACTGCCTCAATTTGCCACTGGAGAAAATCTTCCTCGAGTGGCAGATAT ACTAACGCAACTTTTGCAGACAGATGACTCTGCAGAATTTAACCTAGTGAACAATGCCCTATTAAGTATATTTAAAATGGATGCAAAAGG GACTTTAGGTGGGTTGTTCAGCCAAATACTTCAAGGAGAGGACATTGTTAGAGAACGAGCAATTAAATTCCTTTCTACAAAACTTAAGAC TTTACCAGATGAAGTCTTAACAAAGGAAGTGGAAGAGCTTATACTAACTGAATCCAAAAAGGTCCTAGAAGATGTGACTGGTGAAGAATT TGTTCTATTTATGAAGATACTGTCTGGGTTAAAAAGCTTACAGACAGTGAGTGGAAGACAGCAACTTGTAGAGTTGGTGGCTGAACAGGC CGACCTAGAACAGACCTTCAATCCCTCGGATCCTGACTGTGTGGACAGGCTCTTACAGTGCACTCGGCAGGCAGTACCCCTCTTCTCTAA AAATGTCCATTCCACAAGGTTTGTGACATATTTCTGTGAGCAGGTTCTCCCTAACCTCGGTACCTTGACTACCCCAGTGGAAGGTCTTGA TATACAGTTGGAGGTATTGAAATTGTTGGCGGAGATGAGTTCATTTTGTGGTGACATGGAAAAACTAGAAACAAATTTAAGGAAACTATT TGATAAGTTATTGGAATACATGCCCCTCCCTCCAGAAGAGGCAGAAAATGGAGAGAATGCTGGTAATGAAGAACCCAAGCTACAGTTCAG TTATGTGGAATGTTTGTTGTACAGTTTTCACCAGTTGGGCCGAAAACTTCCAGATTTCTTAACAGCCAAACTGAATGCAGAAAAGCTCAA AGATTTCAAAATCAGGCTGCAGTACTTTGCACGGGGCCTGCAAGTTTATATCAGACAACTTCGCTTAGCTCTCCAGGGTAAAACGGGTGA GGCCTTAAAAACAGAAGAGAACAAGATTAAAGTCGTTGCATTGAAAATAACAAACAATATCAATGTTTTAATCAAGGATCTCTTCCACAT TCCTCCTTCTTATAAGAGCACAGTAACACTATCCTGGAAACCTGTACAAAAGGTTGAGATTGGGCAAAAGAGAGCCAGTGAAGATACAAC TTCAGGTTCACCACCCAAGAAATCTTCAGCAGGACCAAAAAGAGATGCCAGGCAGATTTATAACCCTCCCAGTGGGAAATATAGCAGCAA TTTGGGCAACTTTAATTATGATGGACACATACTGGATTCAAAGAGATATGCCGTTATTGGAGCAGATCTCCGAGACCTGTCTGAACTGGA AGAGAAGCTAAAGAAATGTAACATGAATACACAATTGCCAACACTCCTGATAGCTGAATGTGTGCTGGTTTACATGACTCCAGAGCAGTC CGCAAACCTCCTGAAGTGGGCAGCCAACAGTTTTGAGAGAGCCATGTTCATAAACTACGAACAGGTGAACATGGGTGATCGGTTTGGGCA GATCATGATTGAAAACCTGCGGAGACGCCAGTGTGACCTGGCGGGAGTGGAGACCTGCAAGTCATTAGAGTCACAGAAAGAACGGCTCCT GTCGAATGGGTGGGAAACAGCATCGGCCGTCGACATGATGGAGTTGTACAACAGGTTACCTCGAGCTGAAGTGAGCAGGATAGAATCACT TGAATTCCTGGATGAAATGGAGCTGCTGGAGCAGCTCATGCGGCATTACTGCCTTTGCTGGGCAACCAAAGGAGGAAATGAGCTTGGGCT GAAGGAGATAACTTATTAATCTGTCGAAGGCTTATGCCGAGCCAGAAGCCGAAGCCACTTGCCCTCCTGGAGGAGACCTGCAAGCTCCCT GAGCGGTGGGCGGGCCTCGTCCGCAGGTCTCATCCCACACTCTTGAGAAGCCTTGGTCACTACAGTGGTCGCACATGTTCCTCTTCCTGT TCCTGTTGACATGTCGTTGTTTAAATAAATCTCACTTGCCACCAGTC >5423_5423_2_API5-LCMT1_API5_chr11_43357544_ENST00000420461_LCMT1_chr16_25172422_ENST00000399069_length(amino acids)=628AA_BP=449 MGLSLTMPTVEELYRNYGILADATEQVGQIRRQAIKELPQFATGENLPRVADILTQLLQTDDSAEFNLVNNALLSIFKMDAKGTLGGLFS QILQGEDIVRERAIKFLSTKLKTLPDEVLTKEVEELILTESKKVLEDVTGEEFVLFMKILSGLKSLQTVSGRQQLVELVAEQADLEQTFN PSDPDCVDRLLQCTRQAVPLFSKNVHSTRFVTYFCEQVLPNLGTLTTPVEGLDIQLEVLKLLAEMSSFCGDMEKLETNLRKLFDKLLEYM PLPPEEAENGENAGNEEPKLQFSYVECLLYSFHQLGRKLPDFLTAKLNAEKLKDFKIRLQYFARGLQVYIRQLRLALQGKTGEALKTEEN KIKVVALKITNNINVLIKDLFHIPPSYKSTVTLSWKPVQKVEIGQKRASEDTTSGSPPKKSSAGPKRDARQIYNPPSGKYSSNLGNFNYD GHILDSKRYAVIGADLRDLSELEEKLKKCNMNTQLPTLLIAECVLVYMTPEQSANLLKWAANSFERAMFINYEQVNMGDRFGQIMIENLR RRQCDLAGVETCKSLESQKERLLSNGWETASAVDMMELYNRLPRAEVSRIESLEFLDEMELLEQLMRHYCLCWATKGGNELGLKEITY -------------------------------------------------------------- >5423_5423_3_API5-LCMT1_API5_chr11_43357544_ENST00000455725_LCMT1_chr16_25172422_ENST00000399069_length(transcript)=2451nt_BP=1704nt GCAGCCGCGCTGTGCGCGGTGACTGGCGGCTGCACTGGCGGCAGCTGGAGGTGTAATAGTGCGGGTAGTGGGTTTGGAGAAGTTCCGAGG CGGCGGTGGCGCCGGTCAGGACAAGGATAGCGGAACCGGGCCCTGGGCTTGTCGCTCACCATGCCGACAGTAGAGGAGCTTTACCGCAAT TATGGCATCCTGGCCGATGCCACGGAGCAAGTGGGCCAGACTCATCCATCTCCCCCAACTCAAAAATGAAACAAAGATGGAGGGAGAACG GAATTGGGAAGCATAAAGATGCCTATCAAGTGATATTGGATGGTGTGAAAGGTGGTACTAAGGAAAAGCGATTAGCAGCTCAATTTATTC CGAAATTCTTTAAGCATTTTCCAGAATTGGCTGATTCTGCTATCAATGCACAGTTAGACCTCTGTGAGGATGAAGATGTATCTATTCGAC GTCAAGCAATTAAAGAACTGCCTCAATTTGCCACTGGAGAAAATCTTCCTCGAGTGGCAGATATACTAACGCAACTTTTGCAGACAGATG ACTCTGCAGAATTTAACCTAGTGAACAATGCCCTATTAAGTATATTTAAAATGGATGCAAAAGGGACTTTAGGTGGGTTGTTCAGCCAAA TACTTCAAGGAGAGGACATTGTTAGAGAACGAGCAATTAAATTCCTTTCTACAAAACTTAAGACTTTACCAGATGAAGTCTTAACAAAGG AAGTGGAAGAGCTTATACTAACTGAATCCAAAAAGGTCCTAGAAGATGTGACTGGTGAAGAATTTGTTCTATTTATGAAGATACTGTCTG GGTTAAAAAGCTTACAGACAGTGAGTGGAAGACAGCAACTTGTAGAGTTGGTGGCTGAACAGGCCGACCTAGAACAGACCTTCAATCCCT CGGATCCTGACTGTGTGGACAGGCTCTTACAGTGCACTCGGCAGGCAGTACCCCTCTTCTCTAAAAATGTCCATTCCACAAGGTTTGTGA CATATTTCTGTGAGCAGGTTCTCCCTAACCTCGGTACCTTGACTACCCCAGTGGAAGGTCTTGATATACAGTTGGAGGTATTGAAATTGT TGGCGGAGATGAGTTCATTTTGTGGTGACATGGAAAAACTAGAAACAAATTTAAGGAAACTATTTGATAAGTTATTGGAATACATGCCCC TCCCTCCAGAAGAGGCAGAAAATGGAGAGAATGCTGGTAATGAAGAACCCAAGCTACAGTTCAGTTATGTGGAATGTTTGTTGTACAGTT TTCACCAGTTGGGCCGAAAACTTCCAGATTTCTTAACAGCCAAACTGAATGCAGAAAAGCTCAAAGATTTCAAAATCAGGCTGCAGTACT TTGCACGGGGCCTGCAAGTTTATATCAGACAACTTCGCTTAGCTCTCCAGGGTAAAACGGGTGAGGCCTTAAAAACAGAAGAGAACAAGA TTAAAGTCGTTGCATTGAAAATAACAAACAATATCAATGTTTTAATCAAGGATCTCTTCCACATTCCTCCTTCTTATAAGAGCACAGTAA CACTATCCTGGAAACCTGTACAAAAGGTTGAGATTGGGCAAAAGAGAGCCAGTGAAGATACAACTTCAGGTTCACCACCCAAGAAATCTT CAGCAGGACCAAAAAGAGATGCCAGGCAGATTTATAACCCTCCCAGTGGGAAATATAGCAGCAATTTGGGCAACTTTAATTATGATGGAC ACATACTGGATTCAAAGAGATATGCCGTTATTGGAGCAGATCTCCGAGACCTGTCTGAACTGGAAGAGAAGCTAAAGAAATGTAACATGA ATACACAATTGCCAACACTCCTGATAGCTGAATGTGTGCTGGTTTACATGACTCCAGAGCAGTCCGCAAACCTCCTGAAGTGGGCAGCCA ACAGTTTTGAGAGAGCCATGTTCATAAACTACGAACAGGTGAACATGGGTGATCGGTTTGGGCAGATCATGATTGAAAACCTGCGGAGAC GCCAGTGTGACCTGGCGGGAGTGGAGACCTGCAAGTCATTAGAGTCACAGAAAGAACGGCTCCTGTCGAATGGGTGGGAAACAGCATCGG CCGTCGACATGATGGAGTTGTACAACAGGTTACCTCGAGCTGAAGTGAGCAGGATAGAATCACTTGAATTCCTGGATGAAATGGAGCTGC TGGAGCAGCTCATGCGGCATTACTGCCTTTGCTGGGCAACCAAAGGAGGAAATGAGCTTGGGCTGAAGGAGATAACTTATTAATCTGTCG AAGGCTTATGCCGAGCCAGAAGCCGAAGCCACTTGCCCTCCTGGAGGAGACCTGCAAGCTCCCTGAGCGGTGGGCGGGCCTCGTCCGCAG GTCTCATCCCACACTCTTGAGAAGCCTTGGTCACTACAGTGGTCGCACATGTTCCTCTTCCTGTTCCTGTTGACATGTCGTTGTTTAAAT AAATCTCACTTGCCACCAGTC >5423_5423_3_API5-LCMT1_API5_chr11_43357544_ENST00000455725_LCMT1_chr16_25172422_ENST00000399069_length(amino acids)=665AA_BP=486 MKQRWRENGIGKHKDAYQVILDGVKGGTKEKRLAAQFIPKFFKHFPELADSAINAQLDLCEDEDVSIRRQAIKELPQFATGENLPRVADI LTQLLQTDDSAEFNLVNNALLSIFKMDAKGTLGGLFSQILQGEDIVRERAIKFLSTKLKTLPDEVLTKEVEELILTESKKVLEDVTGEEF VLFMKILSGLKSLQTVSGRQQLVELVAEQADLEQTFNPSDPDCVDRLLQCTRQAVPLFSKNVHSTRFVTYFCEQVLPNLGTLTTPVEGLD IQLEVLKLLAEMSSFCGDMEKLETNLRKLFDKLLEYMPLPPEEAENGENAGNEEPKLQFSYVECLLYSFHQLGRKLPDFLTAKLNAEKLK DFKIRLQYFARGLQVYIRQLRLALQGKTGEALKTEENKIKVVALKITNNINVLIKDLFHIPPSYKSTVTLSWKPVQKVEIGQKRASEDTT SGSPPKKSSAGPKRDARQIYNPPSGKYSSNLGNFNYDGHILDSKRYAVIGADLRDLSELEEKLKKCNMNTQLPTLLIAECVLVYMTPEQS ANLLKWAANSFERAMFINYEQVNMGDRFGQIMIENLRRRQCDLAGVETCKSLESQKERLLSNGWETASAVDMMELYNRLPRAEVSRIESL EFLDEMELLEQLMRHYCLCWATKGGNELGLKEITY -------------------------------------------------------------- >5423_5423_4_API5-LCMT1_API5_chr11_43357544_ENST00000531273_LCMT1_chr16_25172422_ENST00000399069_length(transcript)=2378nt_BP=1631nt GTGCGCGGTGACTGGCGGCTGCACTGGCGGCAGCTGGAGGTGTAATAGTGCGGGTAGTGGGTTTGGAGAAGTTCCGAGGCGGCGGTGGCG CCGGTCAGGACAAGGATAGCGGAACCGGGCCCTGGGCTTGTCGCTCACCATGCCGACAGTAGAGGAGCTTTACCGCAATTATGGCATCCT GGCCGATGCCACGGAGCAAGTGGGCCAGCATAAAGATGCCTATCAAGTGATATTGGATGGTGTGAAAGGTGGTACTAAGGAAAAGCGATT AGCAGCTCAATTTATTCCGAAATTCTTTAAGCATTTTCCAGAATTGGCTGATTCTGCTATCAATGCACAGTTAGACCTCTGTGAGGATGA AGATGTATCTATTCGACGTCAAGCAATTAAAGAACTGCCTCAATTTGCCACTGGAGAAAATCTTCCTCGAGTGGCAGATATACTAACGCA ACTTTTGCAGACAGATGACTCTGCAGAATTTAACCTAGTGAACAATGCCCTATTAAGTATATTTAAAATGGATGCAAAAGGGACTTTAGG TGGGTTGTTCAGCCAAATACTTCAAGGAGAGGACATTGTTAGAGAACGAGCAATTAAATTCCTTTCTACAAAACTTAAGACTTTACCAGA TGAAGTCTTAACAAAGGAAGTGGAAGAGCTTATACTAACTGAATCCAAAAAGGTCCTAGAAGATGTGACTGGTGAAGAATTTGTTCTATT TATGAAGATACTGTCTGGGTTAAAAAGCTTACAGACAGTGAGTGGAAGACAGCAACTTGTAGAGTTGGTGGCTGAACAGGCCGACCTAGA ACAGACCTTCAATCCCTCGGATCCTGACTGTGTGGACAGGCTCTTACAGTGCACTCGGCAGGCAGTACCCCTCTTCTCTAAAAATGTCCA TTCCACAAGGTTTGTGACATATTTCTGTGAGCAGGTTCTCCCTAACCTCGGTACCTTGACTACCCCAGTGGAAGGTCTTGATATACAGTT GGAGGTATTGAAATTGTTGGCGGAGATGAGTTCATTTTGTGGTGACATGGAAAAACTAGAAACAAATTTAAGGAAACTATTTGATAAGTT ATTGGAATACATGCCCCTCCCTCCAGAAGAGGCAGAAAATGGAGAGAATGCTGGTAATGAAGAACCCAAGCTACAGTTCAGTTATGTGGA ATGTTTGTTGTACAGTTTTCACCAGTTGGGCCGAAAACTTCCAGATTTCTTAACAGCCAAACTGAATGCAGAAAAGCTCAAAGATTTCAA AATCAGGCTGCAGTACTTTGCACGGGGCCTGCAAGTTTATATCAGACAACTTCGCTTAGCTCTCCAGGGTAAAACGGGTGAGGCCTTAAA AACAGAAGAGAACAAGATTAAAGTCGTTGCATTGAAAATAACAAACAATATCAATGTTTTAATCAAGGATCTCTTCCACATTCCTCCTTC TTATAAGAGCACAGTAACACTATCCTGGAAACCTGTACAAAAGGTTGAGATTGGGCAAAAGAGAGCCAGTGAAGATACAACTTCAGGTTC ACCACCCAAGAAATCTTCAGCAGGACCAAAAAGAGATGCCAGGCAGATTTATAACCCTCCCAGTGGGAAATATAGCAGCAATTTGGGCAA CTTTAATTATGATGGACACATACTGGATTCAAAGAGATATGCCGTTATTGGAGCAGATCTCCGAGACCTGTCTGAACTGGAAGAGAAGCT AAAGAAATGTAACATGAATACACAATTGCCAACACTCCTGATAGCTGAATGTGTGCTGGTTTACATGACTCCAGAGCAGTCCGCAAACCT CCTGAAGTGGGCAGCCAACAGTTTTGAGAGAGCCATGTTCATAAACTACGAACAGGTGAACATGGGTGATCGGTTTGGGCAGATCATGAT TGAAAACCTGCGGAGACGCCAGTGTGACCTGGCGGGAGTGGAGACCTGCAAGTCATTAGAGTCACAGAAAGAACGGCTCCTGTCGAATGG GTGGGAAACAGCATCGGCCGTCGACATGATGGAGTTGTACAACAGGTTACCTCGAGCTGAAGTGAGCAGGATAGAATCACTTGAATTCCT GGATGAAATGGAGCTGCTGGAGCAGCTCATGCGGCATTACTGCCTTTGCTGGGCAACCAAAGGAGGAAATGAGCTTGGGCTGAAGGAGAT AACTTATTAATCTGTCGAAGGCTTATGCCGAGCCAGAAGCCGAAGCCACTTGCCCTCCTGGAGGAGACCTGCAAGCTCCCTGAGCGGTGG GCGGGCCTCGTCCGCAGGTCTCATCCCACACTCTTGAGAAGCCTTGGTCACTACAGTGGTCGCACATGTTCCTCTTCCTGTTCCTGTTGA CATGTCGTTGTTTAAATAAATCTCACTTGCCACCAGTC >5423_5423_4_API5-LCMT1_API5_chr11_43357544_ENST00000531273_LCMT1_chr16_25172422_ENST00000399069_length(amino acids)=682AA_BP=503 MGLSLTMPTVEELYRNYGILADATEQVGQHKDAYQVILDGVKGGTKEKRLAAQFIPKFFKHFPELADSAINAQLDLCEDEDVSIRRQAIK ELPQFATGENLPRVADILTQLLQTDDSAEFNLVNNALLSIFKMDAKGTLGGLFSQILQGEDIVRERAIKFLSTKLKTLPDEVLTKEVEEL ILTESKKVLEDVTGEEFVLFMKILSGLKSLQTVSGRQQLVELVAEQADLEQTFNPSDPDCVDRLLQCTRQAVPLFSKNVHSTRFVTYFCE QVLPNLGTLTTPVEGLDIQLEVLKLLAEMSSFCGDMEKLETNLRKLFDKLLEYMPLPPEEAENGENAGNEEPKLQFSYVECLLYSFHQLG RKLPDFLTAKLNAEKLKDFKIRLQYFARGLQVYIRQLRLALQGKTGEALKTEENKIKVVALKITNNINVLIKDLFHIPPSYKSTVTLSWK PVQKVEIGQKRASEDTTSGSPPKKSSAGPKRDARQIYNPPSGKYSSNLGNFNYDGHILDSKRYAVIGADLRDLSELEEKLKKCNMNTQLP TLLIAECVLVYMTPEQSANLLKWAANSFERAMFINYEQVNMGDRFGQIMIENLRRRQCDLAGVETCKSLESQKERLLSNGWETASAVDMM ELYNRLPRAEVSRIESLEFLDEMELLEQLMRHYCLCWATKGGNELGLKEITY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for API5-LCMT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for API5-LCMT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for API5-LCMT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | C0012684 | Blastocyst Disintegration | 1 | CTD_human | |
| Tgene | C0013937 | Embryo Resorption | 1 | CTD_human | |
| Tgene | C0752350 | Embryo Death | 1 | CTD_human | |
| Tgene | C0752351 | Embryo Loss | 1 | CTD_human | |
| Tgene | C1136082 | Embryo Disintegration | 1 | CTD_human |
