
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CBFB-PLEKHG4 (FusionGDB2 ID:HG865TG25894) |
Fusion Gene Summary for CBFB-PLEKHG4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CBFB-PLEKHG4 | Fusion gene ID: hg865tg25894 | Hgene | Tgene | Gene symbol | CBFB | PLEKHG4 | Gene ID | 865 | 25894 |
| Gene name | core-binding factor subunit beta | pleckstrin homology and RhoGEF domain containing G4 | |
| Synonyms | PEBP2B | ARHGEF44|PRTPHN1|SCA4 | |
| Cytomap | ('CBFB')('PLEKHG4') 16q22.1 | 16q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | core-binding factor subunit betaCBF-betaPEA2-betaPEBP2-betaSL3-3 enhancer factor 1 beta subunitSL3-3 enhancer factor 1 subunit betaSL3/AKV core-binding factor beta subunitcore-binding factor beta subunitpolyomavirus enhancer binding protein 2, bet | puratrophin-1PH domain-containing family G member 4Purkinje cell atrophy associated protein 1pleckstrin homology domain containing, family G (with RhoGef domain) member 4 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q13951 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000290858, ENST00000412916, ENST00000561924, ENST00000568858, | ||
| Fusion gene scores | * DoF score | 17 X 18 X 12=3672 | 2 X 4 X 2=16 |
| # samples | 33 | 2 | |
| ** MAII score | log2(33/3672*10)=-3.47602812916799 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/16*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: CBFB [Title/Abstract] AND PLEKHG4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CBFB(67116211)-PLEKHG4(67312644), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CBFB-PLEKHG4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CBFB-PLEKHG4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CBFB-PLEKHG4 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. CBFB-PLEKHG4 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CBFB (5'-gene) Fusion gene breakpoints across CBFB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
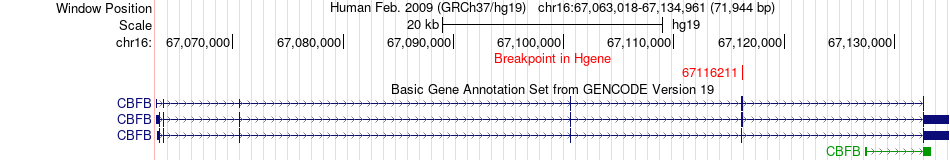 |
 Fusion gene breakpoints across PLEKHG4 (3'-gene) Fusion gene breakpoints across PLEKHG4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
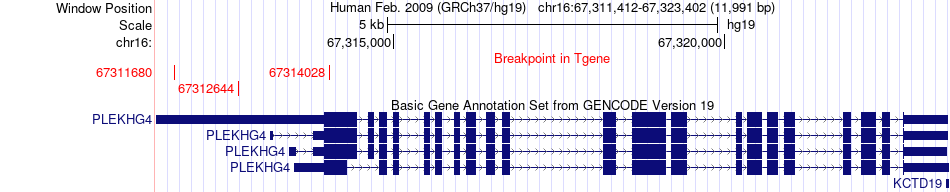 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-1407-01A | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| ChimerDB4 | OV | TCGA-13-1407-01A | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| ChimerDB4 | OV | TCGA-13-1407-01A | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
Top |
Fusion Gene ORF analysis for CBFB-PLEKHG4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000290858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-5UTR | ENST00000290858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-5UTR | ENST00000412916 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-5UTR | ENST00000412916 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-5UTR | ENST00000561924 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-5UTR | ENST00000561924 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000290858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000290858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000290858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000290858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000290858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000290858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000412916 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000412916 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000412916 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000412916 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000412916 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000412916 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000561924 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000561924 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000561924 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000561924 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| 5CDS-intron | ENST00000561924 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| 5CDS-intron | ENST00000561924 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| Frame-shift | ENST00000290858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000290858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000290858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000290858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000412916 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000412916 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000561924 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000561924 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000561924 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| Frame-shift | ENST00000561924 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| In-frame | ENST00000412916 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| In-frame | ENST00000412916 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| intron-3CDS | ENST00000568858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| intron-3CDS | ENST00000568858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| intron-3CDS | ENST00000568858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| intron-3CDS | ENST00000568858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + |
| intron-5UTR | ENST00000568858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| intron-5UTR | ENST00000568858 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| intron-intron | ENST00000568858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| intron-intron | ENST00000568858 | ENST00000379344 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| intron-intron | ENST00000568858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| intron-intron | ENST00000568858 | ENST00000427155 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
| intron-intron | ENST00000568858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67311680 | + |
| intron-intron | ENST00000568858 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67312644 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000412916 | CBFB | chr16 | 67116211 | + | ENST00000360461 | PLEKHG4 | chr16 | 67314028 | + | 4824 | 658 | 163 | 4152 | 1329 |
| ENST00000412916 | CBFB | chr16 | 67116211 | + | ENST00000450733 | PLEKHG4 | chr16 | 67314028 | + | 4601 | 658 | 163 | 3909 | 1248 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000412916 | ENST00000360461 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + | 0.005288378 | 0.99471164 |
| ENST00000412916 | ENST00000450733 | CBFB | chr16 | 67116211 | + | PLEKHG4 | chr16 | 67314028 | + | 0.005479475 | 0.99452055 |
Top |
Fusion Genomic Features for CBFB-PLEKHG4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
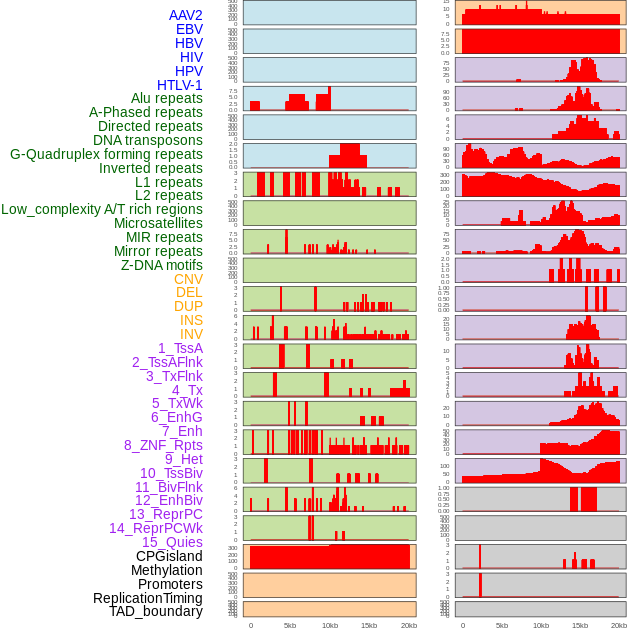 |
Top |
Fusion Protein Features for CBFB-PLEKHG4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:67116211/chr16:67312644) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CBFB | . |
| FUNCTION: Forms the heterodimeric complex core-binding factor (CBF) with RUNX family proteins (RUNX1, RUNX2, and RUNX3). RUNX members modulate the transcription of their target genes through recognizing the core consensus binding sequence 5'-TGTGGT-3', or very rarely, 5'-TGCGGT-3', within their regulatory regions via their runt domain, while CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX. The heterodimers bind to the core site of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, LCK, IL3 and GM-CSF promoters. CBF complexes repress ZBTB7B transcription factor during cytotoxic (CD8+) T cell development. They bind to RUNX-binding sequence within the ZBTB7B locus acting as transcriptional silencer and allowing for cytotoxic T cell differentiation. {ECO:0000250|UniProtKB:Q08024}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000360461 | 0 | 21 | 732_908 | 0 | 1192.0 | Domain | DH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000360461 | 0 | 21 | 920_1027 | 0 | 1192.0 | Domain | PH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000379344 | 0 | 22 | 732_908 | 0 | 1192.0 | Domain | DH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000379344 | 0 | 22 | 920_1027 | 0 | 1192.0 | Domain | PH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000427155 | 0 | 22 | 732_908 | 0 | 1192.0 | Domain | DH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000427155 | 0 | 22 | 920_1027 | 0 | 1192.0 | Domain | PH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000450733 | 0 | 20 | 732_908 | 0 | 1111.0 | Domain | DH | |
| Tgene | PLEKHG4 | chr16:67116211 | chr16:67314028 | ENST00000450733 | 0 | 20 | 920_1027 | 0 | 1111.0 | Domain | PH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for CBFB-PLEKHG4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13376_13376_1_CBFB-PLEKHG4_CBFB_chr16_67116211_ENST00000412916_PLEKHG4_chr16_67314028_ENST00000360461_length(transcript)=4824nt_BP=658nt AGACTCCCCGGAACGGGAGCCCACGCGGGCGGGCGCCTGAAACAAAGGGAAGCGGGCGTCCGGGCGCCGCGGGTGGGCGGTCAGTCGGTC AGCGCGGAGCCAGCCAGCGGGTGCCCGCGCAAGCCCCGAGCGCGGCCGGCCGGCGCGGCCTCAGGGCGGGAAGATGCCGCGCGTCGTGCC CGACCAGAGAAGCAAGTTCGAGAACGAGGAGTTTTTTAGGAAGCTGAGCCGCGAGTGTGAGATTAAGTACACGGGCTTCAGGGACCGGCC CCACGAGGAACGCCAGGCACGCTTCCAGAACGCCTGCCGCGACGGCCGCTCGGAAATCGCTTTTGTGGCCACAGGAACCAATCTGTCTCT CCAGTTTTTTCCGGCCAGCTGGCAGGGAGAACAGCGACAAACACCTAGCCGAGAGTATGTCGACTTAGAAAGAGAAGCAGGCAAGGTATA TTTGAAGGCTCCCATGATTCTGAATGGAGTCTGTGTTATCTGGAAAGGCTGGATTGATCTCCAAAGACTGGATGGTATGGGCTGTCTGGA GTTTGATGAGGAGCGAGCCCAGCAGGAGGATGCATTAGCACAACAGGCCTTTGAAGAGGCTCGGAGAAGGACACGCGAATTTGAAGATAG AGACAGGTCTCATCGGGAGGAAATGGAGTTCAGGGATGCCTGGGAAGAGGAGGAACCTGCTTCCCAGATGCACGTTAAGGACCCAGGTCC TCCAAGACCACCAGCCGGGGCCACCCAGGATGAGGAGCTACAGGGCAGCCCCTTGTCCAGGAAATTCCAGTTACCCCCAGCTGCAGATGA GTCGGGGGATGCCCAGAGGGGCACAGTAGAAAGCTCCTCAGTCCTGTCAGAAGGGCCAGGCCCCTCTGGAGTGGAGAGTCTCCTATGCCC CATGTCCTCCCACCTCAGCTTGGCACAGGGTGAGAGTGACACCCCAGGGGTAGGGTTGGTAGGGGACCCAGGTCCAAGCAGGGCGATGCC ATCTGGCTTGAGCCCTGGGGCATTGGACAGCGACCCTGTGGGCCTTGGAGACCCTTTATCAGAAATATCAAAGCTGCTGGAGGCAGCCCC CAGTGGATCCGGCCTCCCTAAGCCTGCTGACTGCCTCCTGGCCCAAGACCTCTGTTGGGAGCTGCTGGCCAGTGGTATGGCCACCTTGCC AGGGACTCGGGATGTCCAAGGCCGGGCAGTGCTGCTTCTGTGTGCCCACAGCCCAGCCTGGCTTCAGTCTGAGTGCAGCAGCCAGGAACT CATCCGCCTCCTGCTGTACCTGCGAAGCATCCCCAGGCCCGAAGTACAGGCACTGGGACTGACAGTGCTAGTTGATGCCCGAATTTGTGC TCCAAGTTCTTCCCTCTTCTCTGGGCTCAGCCAACTACAAGAAGCAGCCCCAGGGGCCGTGTACCAGGTGCTGCTAGTGGGAAGCACGCT GCTGAAGGAAGTGCCTTCCGGGCTGCAGCTGGAGCAGTTGCCTTCTCAGAGCCTGCTGACCCACATCCCAACGGCGGGGCTGCCCACTTC GCTAGGAGGAGGCCTGCCTTACTGCCACCAGGCCTGGCTGGATTTCCGAAGGCGGCTGGAAGCTCTACTACAGAACTGCCAGGCAGCTTG TGCCCTGCTCCAGGGGGCCATCGAAAGTGTGAAGGCTGTGCCCCAGCCCATGGAGCCTGGGGAGGTCGGTCAGCTGCTACAGCAGACAGA GGTCCTGATGCAGCAGGTGCTAGACTCGCCATGGCTGGCATGGCTACAATGCCAGGGGGGCCGGGAGCTGACATGGCTGAAGCAAGAGGT CCCAGAGGTGACCCTGAGCCCAGACTACAGGACGGCAATGGACAAGGCTGACGAGCTATATGACCGGGTGGATGGATTGCTGCACCAACT GACCCTGCAGAGCAACCAGCGAATACAGGCCCTAGAGTTGGTCCAAACACTGGAGGCCCGGGAAAGCGGACTGCACCAGATTGAAGTGTG GCTGCAGCAGGTGGGCTGGCCAGCACTGGAGGAGGCTGGGGAGCCCTCGCTGGACATGCTGCTCCAGGCCCAAGGCTCTTTTCAGGAGCT GTACCAGGTTGCCCAGGAGCAGGTCAGGCAAGGGGAGAAGTTTCTGCAGCCGCTGACTGGCTGGGAGGCGGCTGAACTGGACCCCCCTGG GGCACGCTTTCTGGCCCTGCGAGCCCAGCTGACTGAATTCTCTAGGGCTTTGGCCCAGCGGTGCCAGCGGCTGGCGGATGCTGAGAGGCT GTTTCAGCTCTTCAGGGAGGCCTTGACGTGGGCTGAGGAGGGGCAGCGAGTGTTGGCAGAGCTGGAGCAGGAACGCCCGGGGGTTGTGTT GCAGCAGCTGCAGCTGCACTGGACCAGGCACCCTGACTTGCCTCCTGCCCACTTCCGAAAGATGTGGGCTCTGGCCACGGGGCTGGGCTC AGAGGCCATCCGCCAGGAGTGCCGCTGGGCCTGGGCGCGGTGCCAGGACACCTGGCTGGCCCTGGACCAAAAGCTTGAGGCTTCACTGAA GCTACCACCGGTGGGCAGCACAGCTAGCCTGTGTGTCAGCCAGGTCCCCGCTGCACCTGCCCACCCTCCCCTGAGGAAGGCCTACAGCTT CGATCGGAATCTGGGGCAGAGTCTCAGTGAACCTGCCTGCCACTGCCACCATGCGGCCACTATTGCTGCCTGCCGCAGACCAGAGGCTGG AGGAGGTGCCCTGCCCCAGGCATCCCCTACTGTGCCTCCACCAGGCAGCTCTGACCCCAGGAGCCTCAACAGGCTACAGCTGGTGCTGGC AGAGATGGTGGCCACGGAGCGGGAGTATGTCCGGGCTCTAGAGTACACTATGGAGAACTATTTCCCCGAGCTGGATCGCCCCGATGTGCC CCAGGGCCTCCGCGGTCAGCGTGCCCACCTCTTTGGCAACCTGGAGAAGCTGCGGGACTTCCACTGCCACTTCTTCCTGCGTGAGCTGGA GGCTTGCACCCGGCACCCACCACGAGTGGCCTATGCCTTCCTGCGCCATAGGGTGCAGTTTGGGATGTACGCGCTCTACAGCAAGAATAA GCCTCGCTCCGATGCCCTGATGTCAAGCTATGGGCACACCTTCTTCAAGGACAAGCAGCAAGCACTGGGGGACCACCTGGACCTGGCCTC CTACCTGCTAAAGCCCATCCAGCGCATGGGCAAGTACGCACTGCTGCTGCAGGAGCTGGCACGGGCCTGCGGGGGCCCCACGCAGGAGCT CAGTGCGCTGCGGGAGGCCCAGAGCCTTGTGCACTTCCAGCTGCGGCACGGAAACGACCTGCTGGCCATGGACGCCATCCAGGGCTGTGA TGTTAACCTCAAGGAACAGGGGCAGCTGGTGCGACAGGATGAGTTTGTGGTGCGCACTGGGCGCCACAAGTCCGTGCGCCGCATCTTCCT TTTTGAGGAGCTGCTGCTCTTCAGCAAGCCTCGCCATGGGCCCACAGGGGTTGACACATTTGCCTACAAGCGCTCCTTCAAGATGGCAGA CCTTGGTCTCACTGAGTGCTGTGGGAACAGCAACCTGCGCTTCGAGATCTGGTTCCGCCGCCGCAAGGCCAGGGACACCTTTGTGCTGCA GGCCTCCAGCCTGGCTATCAAGCAGGCCTGGACAGCTGACATCTCCCACCTGCTTTGGAGGCAGGCCGTCCACAACAAGGAGGTGCGCAT GGCTGAGATGGTGTCCATGGGTGTGGGGAACAAGGCCTTCCGAGACATTGCTCCCAGCGAGGAAGCCATCAACGACCGCACCGTCAACTA TGTCCTGAAGTGCCGAGAAGTTCGCTCTCGGGCGTCCATTGCCGTAGCCCCGTTTGACCATGACAGCCTCTACCTGGGGGCCTCGAACTC CCTTCCTGGAGACCCTGCCTCTTGCTCTGTTCTGGGGTCCCTCAACCTGCACCTGTACAGAGACCCAGCTCTTCTGGGTCTCCGCTGTCC CCTGTATCCCAGCTTCCCAGAGGAAGCAGCACTGGAGGCTGAGGCAGAGCTGGGCGGCCAGCCCTCTTTGACTGCTGAGGACTCAGAGAT CTCGTCCCAATGCCCATCAGCCAGTGGCTCCAGTGGCTCTGACAGCAGCTGTGTGTCAGGGCAGGCCCTGGGTAGGGGCCTGGAGGACTT ACCCTGTGTCTGAGCCCGGGACTGGACGAGCAGTAGATCCAGCAGCCTGCAGCTCCAAGGAACATTGCCTCTCTGGATCTGCTGTGACCA GGGTGTGGCTGACACCTGGGCTACCTCCAACCTACATGTGCAACGCTGTTGACTACCCTTTCTGATGTGTGTGGCCATTGGACTAACTGG CACGGGGCCTCTCTAGGGAAGTCTGGTTGTAGAGCCTGAATAGGCTCCTGGCCCCATGACCCCTTCTCCTGTCCCCAGCTCCCATCCCAG TTGTGGGTTAAGAATAGGCTAGAGCAGACATTGGGTGTTTCCATGCTGTAGGCTGGTGGGGGACCATGTGCCTCTAGGCAGTGACTAGGG TGCCCCCACCCCTCAGGAAGAACACAGGTGGGCTCCTAGCAGCTGATCCCCAATGCCTGGCCTTAAAGCCGAGCTCAGTTACCATAGGGA CAGGTCCACCTCTACTGGGCCCTCATGCTTGCCTTTCCTGGCCCCCAGGCCCAGCCCCTTTTTACTGGGGCAGTTTCGTTATTTTGACTT GATGCCTTTTGAATAACTTTCAATAGAATTGTCTAAAATTATCTTACTGGTTGTTAGGCCTTTGGTGTCTCAGAGAAGGAGTCTAGGTCT TTGATGTGTGATTTAATCTTTTATTTGTTTATAATAAAAAATAGACTGATATGT >13376_13376_1_CBFB-PLEKHG4_CBFB_chr16_67116211_ENST00000412916_PLEKHG4_chr16_67314028_ENST00000360461_length(amino acids)=1329AA_BP=0 MPRVVPDQRSKFENEEFFRKLSRECEIKYTGFRDRPHEERQARFQNACRDGRSEIAFVATGTNLSLQFFPASWQGEQRQTPSREYVDLER EAGKVYLKAPMILNGVCVIWKGWIDLQRLDGMGCLEFDEERAQQEDALAQQAFEEARRRTREFEDRDRSHREEMEFRDAWEEEEPASQMH VKDPGPPRPPAGATQDEELQGSPLSRKFQLPPAADESGDAQRGTVESSSVLSEGPGPSGVESLLCPMSSHLSLAQGESDTPGVGLVGDPG PSRAMPSGLSPGALDSDPVGLGDPLSEISKLLEAAPSGSGLPKPADCLLAQDLCWELLASGMATLPGTRDVQGRAVLLLCAHSPAWLQSE CSSQELIRLLLYLRSIPRPEVQALGLTVLVDARICAPSSSLFSGLSQLQEAAPGAVYQVLLVGSTLLKEVPSGLQLEQLPSQSLLTHIPT AGLPTSLGGGLPYCHQAWLDFRRRLEALLQNCQAACALLQGAIESVKAVPQPMEPGEVGQLLQQTEVLMQQVLDSPWLAWLQCQGGRELT WLKQEVPEVTLSPDYRTAMDKADELYDRVDGLLHQLTLQSNQRIQALELVQTLEARESGLHQIEVWLQQVGWPALEEAGEPSLDMLLQAQ GSFQELYQVAQEQVRQGEKFLQPLTGWEAAELDPPGARFLALRAQLTEFSRALAQRCQRLADAERLFQLFREALTWAEEGQRVLAELEQE RPGVVLQQLQLHWTRHPDLPPAHFRKMWALATGLGSEAIRQECRWAWARCQDTWLALDQKLEASLKLPPVGSTASLCVSQVPAAPAHPPL RKAYSFDRNLGQSLSEPACHCHHAATIAACRRPEAGGGALPQASPTVPPPGSSDPRSLNRLQLVLAEMVATEREYVRALEYTMENYFPEL DRPDVPQGLRGQRAHLFGNLEKLRDFHCHFFLRELEACTRHPPRVAYAFLRHRVQFGMYALYSKNKPRSDALMSSYGHTFFKDKQQALGD HLDLASYLLKPIQRMGKYALLLQELARACGGPTQELSALREAQSLVHFQLRHGNDLLAMDAIQGCDVNLKEQGQLVRQDEFVVRTGRHKS VRRIFLFEELLLFSKPRHGPTGVDTFAYKRSFKMADLGLTECCGNSNLRFEIWFRRRKARDTFVLQASSLAIKQAWTADISHLLWRQAVH NKEVRMAEMVSMGVGNKAFRDIAPSEEAINDRTVNYVLKCREVRSRASIAVAPFDHDSLYLGASNSLPGDPASCSVLGSLNLHLYRDPAL LGLRCPLYPSFPEEAALEAEAELGGQPSLTAEDSEISSQCPSASGSSGSDSSCVSGQALGRGLEDLPCV -------------------------------------------------------------- >13376_13376_2_CBFB-PLEKHG4_CBFB_chr16_67116211_ENST00000412916_PLEKHG4_chr16_67314028_ENST00000450733_length(transcript)=4601nt_BP=658nt AGACTCCCCGGAACGGGAGCCCACGCGGGCGGGCGCCTGAAACAAAGGGAAGCGGGCGTCCGGGCGCCGCGGGTGGGCGGTCAGTCGGTC AGCGCGGAGCCAGCCAGCGGGTGCCCGCGCAAGCCCCGAGCGCGGCCGGCCGGCGCGGCCTCAGGGCGGGAAGATGCCGCGCGTCGTGCC CGACCAGAGAAGCAAGTTCGAGAACGAGGAGTTTTTTAGGAAGCTGAGCCGCGAGTGTGAGATTAAGTACACGGGCTTCAGGGACCGGCC CCACGAGGAACGCCAGGCACGCTTCCAGAACGCCTGCCGCGACGGCCGCTCGGAAATCGCTTTTGTGGCCACAGGAACCAATCTGTCTCT CCAGTTTTTTCCGGCCAGCTGGCAGGGAGAACAGCGACAAACACCTAGCCGAGAGTATGTCGACTTAGAAAGAGAAGCAGGCAAGGTATA TTTGAAGGCTCCCATGATTCTGAATGGAGTCTGTGTTATCTGGAAAGGCTGGATTGATCTCCAAAGACTGGATGGTATGGGCTGTCTGGA GTTTGATGAGGAGCGAGCCCAGCAGGAGGATGCATTAGCACAACAGGCCTTTGAAGAGGCTCGGAGAAGGACACGCGAATTTGAAGATAG AGACAGGTCTCATCGGGAGGAAATGGAGTTCAGGGATGCCTGGGAAGAGGAGGAACCTGCTTCCCAGATGCACGTTAAGGACCCAGGTCC TCCAAGACCACCAGCCGGGGCCACCCAGGATGAGGAGCTACAGGGCAGCCCCTTGTCCAGGAAATTCCAGTTACCCCCAGCTGCAGATGA GTCGGGGGATGCCCAGAGGGGCACAGTAGAAAGCTCCTCAGTCCTGTCAGAAGGGCCAGGCCCCTCTGGAGTGGAGAGTCTCCTATGCCC CATGTCCTCCCACCTCAGCTTGGCACAGGGGACTCGGGATGTCCAAGGCCGGGCAGTGCTGCTTCTGTGTGCCCACAGCCCAGCCTGGCT TCAGTCTGAGTGCAGCAGCCAGGAACTCATCCGCCTCCTGCTGTACCTGCGAAGCATCCCCAGGCCCGAAGTACAGGCACTGGGACTGAC AGTGCTAGTTGATGCCCGAATTTGTGCTCCAAGTTCTTCCCTCTTCTCTGGGCTCAGCCAACTACAAGAAGCAGCCCCAGGGGCCGTGTA CCAGGTGCTGCTAGTGGGAAGCACGCTGCTGAAGGAAGTGCCTTCCGGGCTGCAGCTGGAGCAGTTGCCTTCTCAGAGCCTGCTGACCCA CATCCCAACGGCGGGGCTGCCCACTTCGCTAGGAGGAGGCCTGCCTTACTGCCACCAGGCCTGGCTGGATTTCCGAAGGCGGCTGGAAGC TCTACTACAGAACTGCCAGGCAGCTTGTGCCCTGCTCCAGGGGGCCATCGAAAGTGTGAAGGCTGTGCCCCAGCCCATGGAGCCTGGGGA GGTCGGTCAGCTGCTACAGCAGACAGAGGTCCTGATGCAGCAGGTGCTAGACTCGCCATGGCTGGCATGGCTACAATGCCAGGGGGGCCG GGAGCTGACATGGCTGAAGCAAGAGGTCCCAGAGGTGACCCTGAGCCCAGACTACAGGACGGCAATGGACAAGGCTGACGAGCTATATGA CCGGGTGGATGGATTGCTGCACCAACTGACCCTGCAGAGCAACCAGCGAATACAGGCCCTAGAGTTGGTCCAAACACTGGAGGCCCGGGA AAGCGGACTGCACCAGATTGAAGTGTGGCTGCAGCAGGTGGGCTGGCCAGCACTGGAGGAGGCTGGGGAGCCCTCGCTGGACATGCTGCT CCAGGCCCAAGGCTCTTTTCAGGAGCTGTACCAGGTTGCCCAGGAGCAGGTCAGGCAAGGGGAGAAGTTTCTGCAGCCGCTGACTGGCTG GGAGGCGGCTGAACTGGACCCCCCTGGGGCACGCTTTCTGGCCCTGCGAGCCCAGCTGACTGAATTCTCTAGGGCTTTGGCCCAGCGGTG CCAGCGGCTGGCGGATGCTGAGAGGCTGTTTCAGCTCTTCAGGGAGGCCTTGACGTGGGCTGAGGAGGGGCAGCGAGTGTTGGCAGAGCT GGAGCAGGAACGCCCGGGGGTTGTGTTGCAGCAGCTGCAGCTGCACTGGACCAGGCACCCTGACTTGCCTCCTGCCCACTTCCGAAAGAT GTGGGCTCTGGCCACGGGGCTGGGCTCAGAGGCCATCCGCCAGGAGTGCCGCTGGGCCTGGGCGCGGTGCCAGGACACCTGGCTGGCCCT GGACCAAAAGCTTGAGGCTTCACTGAAGCTACCACCGGTGGGCAGCACAGCTAGCCTGTGTGTCAGCCAGGTCCCCGCTGCACCTGCCCA CCCTCCCCTGAGGAAGGCCTACAGCTTCGATCGGAATCTGGGGCAGAGTCTCAGTGAACCTGCCTGCCACTGCCACCATGCGGCCACTAT TGCTGCCTGCCGCAGACCAGAGGCTGGAGGAGGTGCCCTGCCCCAGGCATCCCCTACTGTGCCTCCACCAGGCAGCTCTGACCCCAGGAG CCTCAACAGGCTACAGCTGGTGCTGGCAGAGATGGTGGCCACGGAGCGGGAGTATGTCCGGGCTCTAGAGTACACTATGGAGAACTATTT CCCCGAGCTGGATCGCCCCGATGTGCCCCAGGGCCTCCGCGGTCAGCGTGCCCACCTCTTTGGCAACCTGGAGAAGCTGCGGGACTTCCA CTGCCACTTCTTCCTGCGTGAGCTGGAGGCTTGCACCCGGCACCCACCACGAGTGGCCTATGCCTTCCTGCGCCATAGGGTGCAGTTTGG GATGTACGCGCTCTACAGCAAGAATAAGCCTCGCTCCGATGCCCTGATGTCAAGCTATGGGCACACCTTCTTCAAGGACAAGCAGCAAGC ACTGGGGGACCACCTGGACCTGGCCTCCTACCTGCTAAAGCCCATCCAGCGCATGGGCAAGTACGCACTGCTGCTGCAGGAGCTGGCACG GGCCTGCGGGGGCCCCACGCAGGAGCTCAGTGCGCTGCGGGAGGCCCAGAGCCTTGTGCACTTCCAGCTGCGGCACGGAAACGACCTGCT GGCCATGGACGCCATCCAGGGCTGTGATGTTAACCTCAAGGAACAGGGGCAGCTGGTGCGACAGGATGAGTTTGTGGTGCGCACTGGGCG CCACAAGTCCGTGCGCCGCATCTTCCTTTTTGAGGAGCTGCTGCTCTTCAGCAAGCCTCGCCATGGGCCCACAGGGGTTGACACATTTGC CTACAAGCGCTCCTTCAAGATGGCAGACCTTGGTCTCACTGAGTGCTGTGGGAACAGCAACCTGCGCTTCGAGATCTGGTTCCGCCGCCG CAAGGCCAGGGACACCTTTGTGCTGCAGGCCTCCAGCCTGGCTATCAAGCAGGCCTGGACAGCTGACATCTCCCACCTGCTTTGGAGGCA GGCCGTCCACAACAAGGAGGTGCGCATGGCTGAGATGGTGTCCATGGGTGTGGGGAACAAGGCCTTCCGAGACATTGCTCCCAGCGAGGA AGCCATCAACGACCGCACCGTCAACTATGTCCTGAAGTGCCGAGAAGTTCGCTCTCGGGCGTCCATTGCCGTAGCCCCGTTTGACCATGA CAGCCTCTACCTGGGGGCCTCGAACTCCCTTCCTGGAGACCCTGCCTCTTGCTCTGTTCTGGGGTCCCTCAACCTGCACCTGTACAGAGA CCCAGCTCTTCTGGGTCTCCGCTGTCCCCTGTATCCCAGCTTCCCAGAGGAAGCAGCACTGGAGGCTGAGGCAGAGCTGGGCGGCCAGCC CTCTTTGACTGCTGAGGACTCAGAGATCTCGTCCCAATGCCCATCAGCCAGTGGCTCCAGTGGCTCTGACAGCAGCTGTGTGTCAGGGCA GGCCCTGGGTAGGGGCCTGGAGGACTTACCCTGTGTCTGAGCCCGGGACTGGACGAGCAGTAGATCCAGCAGCCTGCAGCTCCAAGGAAC ATTGCCTCTCTGGATCTGCTGTGACCAGGGTGTGGCTGACACCTGGGCTACCTCCAACCTACATGTGCAACGCTGTTGACTACCCTTTCT GATGTGTGTGGCCATTGGACTAACTGGCACGGGGCCTCTCTAGGGAAGTCTGGTTGTAGAGCCTGAATAGGCTCCTGGCCCCATGACCCC TTCTCCTGTCCCCAGCTCCCATCCCAGTTGTGGGTTAAGAATAGGCTAGAGCAGACATTGGGTGTTTCCATGCTGTAGGCTGGTGGGGGA CCATGTGCCTCTAGGCAGTGACTAGGGTGCCCCCACCCCTCAGGAAGAACACAGGTGGGCTCCTAGCAGCTGATCCCCAATGCCTGGCCT TAAAGCCGAGCTCAGTTACCATAGGGACAGGTCCACCTCTACTGGGCCCTCATGCTTGCCTTTCCTGGCCCCCAGGCCCAGCCCCTTTTT ACTGGGGCAGTTTCGTTATTTTGACTTGATGCCTTTTGAATAACTTTCAATAGAATTGTCTAAAATTATCTTACTGGTTGTTAGGCCTTT GGTGTCTCAGAGAAGGAGTCTAGGTCTTTGATGTGTGATTTAATCTTTTATTTGTTTATAATAAAAAATAGACTGATATGTACCCTCTGA TGGGCACATGC >13376_13376_2_CBFB-PLEKHG4_CBFB_chr16_67116211_ENST00000412916_PLEKHG4_chr16_67314028_ENST00000450733_length(amino acids)=1248AA_BP=0 MPRVVPDQRSKFENEEFFRKLSRECEIKYTGFRDRPHEERQARFQNACRDGRSEIAFVATGTNLSLQFFPASWQGEQRQTPSREYVDLER EAGKVYLKAPMILNGVCVIWKGWIDLQRLDGMGCLEFDEERAQQEDALAQQAFEEARRRTREFEDRDRSHREEMEFRDAWEEEEPASQMH VKDPGPPRPPAGATQDEELQGSPLSRKFQLPPAADESGDAQRGTVESSSVLSEGPGPSGVESLLCPMSSHLSLAQGTRDVQGRAVLLLCA HSPAWLQSECSSQELIRLLLYLRSIPRPEVQALGLTVLVDARICAPSSSLFSGLSQLQEAAPGAVYQVLLVGSTLLKEVPSGLQLEQLPS QSLLTHIPTAGLPTSLGGGLPYCHQAWLDFRRRLEALLQNCQAACALLQGAIESVKAVPQPMEPGEVGQLLQQTEVLMQQVLDSPWLAWL QCQGGRELTWLKQEVPEVTLSPDYRTAMDKADELYDRVDGLLHQLTLQSNQRIQALELVQTLEARESGLHQIEVWLQQVGWPALEEAGEP SLDMLLQAQGSFQELYQVAQEQVRQGEKFLQPLTGWEAAELDPPGARFLALRAQLTEFSRALAQRCQRLADAERLFQLFREALTWAEEGQ RVLAELEQERPGVVLQQLQLHWTRHPDLPPAHFRKMWALATGLGSEAIRQECRWAWARCQDTWLALDQKLEASLKLPPVGSTASLCVSQV PAAPAHPPLRKAYSFDRNLGQSLSEPACHCHHAATIAACRRPEAGGGALPQASPTVPPPGSSDPRSLNRLQLVLAEMVATEREYVRALEY TMENYFPELDRPDVPQGLRGQRAHLFGNLEKLRDFHCHFFLRELEACTRHPPRVAYAFLRHRVQFGMYALYSKNKPRSDALMSSYGHTFF KDKQQALGDHLDLASYLLKPIQRMGKYALLLQELARACGGPTQELSALREAQSLVHFQLRHGNDLLAMDAIQGCDVNLKEQGQLVRQDEF VVRTGRHKSVRRIFLFEELLLFSKPRHGPTGVDTFAYKRSFKMADLGLTECCGNSNLRFEIWFRRRKARDTFVLQASSLAIKQAWTADIS HLLWRQAVHNKEVRMAEMVSMGVGNKAFRDIAPSEEAINDRTVNYVLKCREVRSRASIAVAPFDHDSLYLGASNSLPGDPASCSVLGSLN LHLYRDPALLGLRCPLYPSFPEEAALEAEAELGGQPSLTAEDSEISSQCPSASGSSGSDSSCVSGQALGRGLEDLPCV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CBFB-PLEKHG4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CBFB-PLEKHG4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CBFB-PLEKHG4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CBFB | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Hgene | CBFB | C0023479 | Acute myelomonocytic leukemia | 2 | CTD_human;ORPHANET |
| Hgene | CBFB | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Hgene | CBFB | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Hgene | CBFB | C0005941 | Bone Diseases, Developmental | 1 | CTD_human |
| Hgene | CBFB | C0008925 | Cleft Palate | 1 | CTD_human |
| Hgene | CBFB | C0018798 | Congenital Heart Defects | 1 | CTD_human |
| Hgene | CBFB | C0029396 | Heterotopic Ossification | 1 | CTD_human |
| Hgene | CBFB | C1837218 | Cleft palate, isolated | 1 | CTD_human |
| Tgene | C0752122 | Spinocerebellar Ataxia Type 4 | 2 | ORPHANET |
