
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CBL-CXCR5 (FusionGDB2 ID:HG867TG643) |
Fusion Gene Summary for CBL-CXCR5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CBL-CXCR5 | Fusion gene ID: hg867tg643 | Hgene | Tgene | Gene symbol | CBL | CXCR5 | Gene ID | 867 | 643 |
| Gene name | Cbl proto-oncogene | C-X-C motif chemokine receptor 5 | |
| Synonyms | C-CBL|CBL2|FRA11B|NSLL|RNF55 | BLR1|CD185|MDR15 | |
| Cytomap | ('CBL')('CXCR5') 11q23.3 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase CBLCas-Br-M (murine) ecotropic retroviral transforming sequenceCbl proto-oncogene, E3 ubiquitin protein ligaseRING finger protein 55RING-type E3 ubiquitin transferase CBLcasitas B-lineage lymphoma proto-oncogenefragile si | C-X-C chemokine receptor type 5Burkitt lymphoma receptor 1, GTP binding protein (chemokine (C-X-C motif) receptor 5)Burkitt lymphoma receptor 1, GTP-binding proteinCXC-R5CXCR-5MDR-15chemokine (C-X-C motif) receptor 5monocyte-derived receptor 15 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P22681 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000264033, | ||
| Fusion gene scores | * DoF score | 13 X 8 X 8=832 | 12 X 4 X 8=384 |
| # samples | 18 | 12 | |
| ** MAII score | log2(18/832*10)=-2.20858662181142 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/384*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: CBL [Title/Abstract] AND CXCR5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CBL(119103405)-CXCR5(118764305), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CBL-CXCR5 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CBL-CXCR5 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CBL (5'-gene) Fusion gene breakpoints across CBL (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
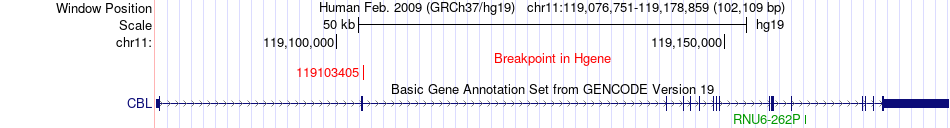 |
 Fusion gene breakpoints across CXCR5 (3'-gene) Fusion gene breakpoints across CXCR5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
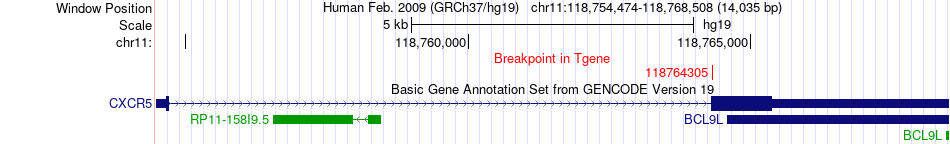 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-J9-A8CL-01A | CBL | chr11 | 119103405 | - | CXCR5 | chr11 | 118764305 | + |
| ChimerDB4 | PRAD | TCGA-J9-A8CL-01A | CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764305 | + |
| ChimerDB4 | PRAD | TCGA-J9-A8CL | CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764305 | + |
Top |
Fusion Gene ORF analysis for CBL-CXCR5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000264033 | ENST00000292174 | CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764305 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000264033 | CBL | chr11 | 119103405 | + | ENST00000292174 | CXCR5 | chr11 | 118764305 | + | 5023 | 819 | 828 | 1886 | 352 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000264033 | ENST00000292174 | CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764305 | + | 0.086296104 | 0.9137039 |
Top |
Fusion Genomic Features for CBL-CXCR5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764304 | + | 5.13E-06 | 0.9999949 |
| CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764304 | + | 5.13E-06 | 0.9999949 |
| CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764304 | + | 5.13E-06 | 0.9999949 |
| CBL | chr11 | 119103405 | + | CXCR5 | chr11 | 118764304 | + | 5.13E-06 | 0.9999949 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
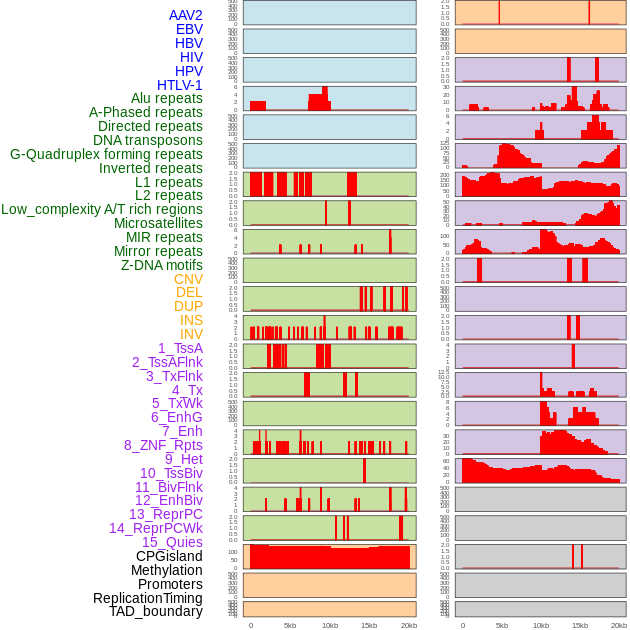 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
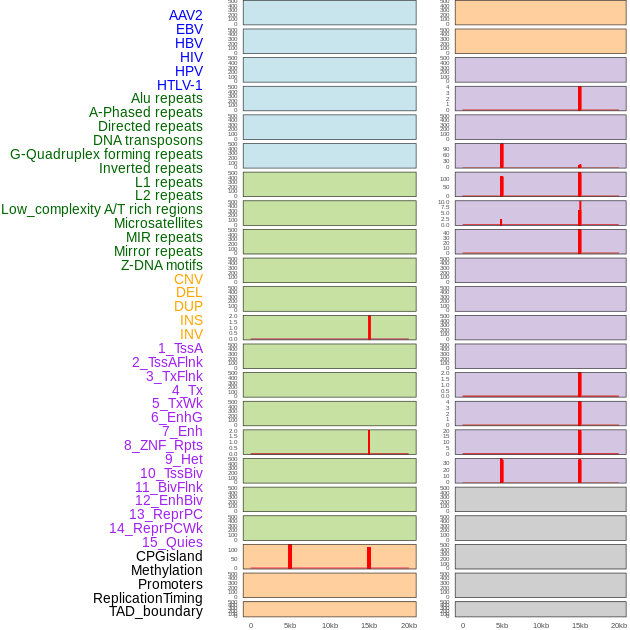 |
Top |
Fusion Protein Features for CBL-CXCR5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:119103405/chr11:118764305) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CBL | . |
| FUNCTION: Adapter protein that functions as a negative regulator of many signaling pathways that are triggered by activation of cell surface receptors. Acts as an E3 ubiquitin-protein ligase, which accepts ubiquitin from specific E2 ubiquitin-conjugating enzymes, and then transfers it to substrates promoting their degradation by the proteasome (PubMed:17094949). Ubiquitinates SPRY2 (PubMed:17094949, PubMed:17974561). Ubiquitinates EGFR (PubMed:17974561). Recognizes activated receptor tyrosine kinases, including KIT, FLT1, FGFR1, FGFR2, PDGFRA, PDGFRB, CSF1R, EPHA8 and KDR and terminates signaling. Recognizes membrane-bound HCK, SRC and other kinases of the SRC family and mediates their ubiquitination and degradation. Participates in signal transduction in hematopoietic cells. Plays an important role in the regulation of osteoblast differentiation and apoptosis. Essential for osteoclastic bone resorption. The 'Tyr-731' phosphorylated form induces the activation and recruitment of phosphatidylinositol 3-kinase to the cell membrane in a signaling pathway that is critical for osteoclast function. May be functionally coupled with the E2 ubiquitin-protein ligase UB2D3. In association with CBLB, required for proper feedback inhibition of ciliary platelet-derived growth factor receptor-alpha (PDGFRA) signaling pathway via ubiquitination and internalization of PDGFRA (By similarity). {ECO:0000250|UniProtKB:P22682, ECO:0000269|PubMed:10514377, ECO:0000269|PubMed:11896602, ECO:0000269|PubMed:14661060, ECO:0000269|PubMed:14739300, ECO:0000269|PubMed:15190072, ECO:0000269|PubMed:17094949, ECO:0000269|PubMed:17509076, ECO:0000269|PubMed:17974561, ECO:0000269|PubMed:18374639, ECO:0000269|PubMed:19689429, ECO:0000269|PubMed:21596750}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 110_124 | 17 | 373.0 | Topological domain | Extracellular | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 146_167 | 17 | 373.0 | Topological domain | Cytoplasmic | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 189_219 | 17 | 373.0 | Topological domain | Extracellular | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 241_259 | 17 | 373.0 | Topological domain | Cytoplasmic | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 281_304 | 17 | 373.0 | Topological domain | Extracellular | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 326_372 | 17 | 373.0 | Topological domain | Cytoplasmic | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 77_88 | 17 | 373.0 | Topological domain | Cytoplasmic | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 125_145 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 168_188 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 220_240 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 260_280 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 305_325 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D7 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 56_76 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 89_109 | 17 | 373.0 | Transmembrane | Helical%3B Name%3D2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 227_240 | 147 | 907.0 | Calcium binding | . |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 357_476 | 147 | 907.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 477_688 | 147 | 907.0 | Compositional bias | Note=Pro-rich |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 689_834 | 147 | 907.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 47_351 | 147 | 907.0 | Domain | Cbl-PTB |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 856_895 | 147 | 907.0 | Domain | UBA |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 176_248 | 147 | 907.0 | Region | Note=EF-hand-like |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 249_351 | 147 | 907.0 | Region | Note=SH2-like |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 352_380 | 147 | 907.0 | Region | Note=Linker |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 358_906 | 147 | 907.0 | Region | Required for ubiquitination of SPRED2 |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 47_175 | 147 | 907.0 | Region | Note=4H |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 381_420 | 147 | 907.0 | Zinc finger | RING-type |
| Tgene | CXCR5 | chr11:119103405 | chr11:118764305 | ENST00000292174 | 0 | 2 | 1_55 | 17 | 373.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for CBL-CXCR5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >13396_13396_1_CBL-CXCR5_CBL_chr11_119103405_ENST00000264033_CXCR5_chr11_118764305_ENST00000292174_length(transcript)=5023nt_BP=819nt ACAGAAACCTGGCCGCGGCTTTCCGGGAGCTGTGGGCACGCGAGCTGCTCGAAGCCGGTGGCCCGGGGATCGTCGCTGCCCAGTCCGGCC GCGGCCGGCCGCTTCCCCGAGCTTACCAGTAAGCCGGTCCGCCTCCGGGACGCGCCCCGCCCGCCCCGGGCTCGCGCGCTCTGAAGGGGC GCGTCCCGCGCACTCCCGGCGCCGCCCACTCCCCTCCCCACGGCCGCTCCTCCCTCCGCCCGGATAGCCGGCGGCGGCGGCGGCGGCGGC GGCGGCGGCGGCCGGGAGAGGCCCCTCCTTCACGCCCTGCTTCTCTCCCTCGCTCGCAGTCGAGCCGAGCCGGCGGACCCGCCTGGGCTC CGACCCTGCCCAGGCCATGGCCGGCAACGTGAAGAAGAGCTCTGGGGCCGGGGGCGGCAGCGGCTCCGGGGGCTCGGGTTCGGGTGGCCT GATTGGGCTCATGAAGGACGCCTTCCAGCCGCACCACCACCACCACCACCACCTCAGCCCCCACCCGCCGGGGACGGTGGACAAGAAGAT GGTGGAGAAGTGCTGGAAGCTCATGGACAAGGTGGTGCGGTTGTGTCAGAACCCAAAGCTGGCGCTAAAGAATAGCCCACCTTATATCTT AGACCTGCTACCAGATACCTACCAGCATCTCCGTACTATCTTGTCAAGATATGAGGGGAAGATGGAGACACTTGGAGAAAATGAGTATTT TAGGGTGTTTATGGAGAATTTGATGAAGAAAACTAAGCAAACCATAAGCCTCTTCAAGGAGGGAAAAGAAAGAATGTATGAGGAGAATTC TCAGCCTAGTTCTGGGAACTGGACAGATTGGACAACTATAACGACACCTCCCTGGTGGAAAATCATCTCTGCCCTGCCACAGAGGGGCCC CTCATGGCCTCCTTCAAGGCCGTGTTCGTGCCCGTGGCCTACAGCCTCATCTTCCTCCTGGGCGTGATCGGCAACGTCCTGGTGCTGGTG ATCCTGGAGCGGCACCGGCAGACACGCAGTTCCACGGAGACCTTCCTGTTCCACCTGGCCGTGGCCGACCTCCTGCTGGTCTTCATCTTG CCCTTTGCCGTGGCCGAGGGCTCTGTGGGCTGGGTCCTGGGGACCTTCCTCTGCAAAACTGTGATTGCCCTGCACAAAGTCAACTTCTAC TGCAGCAGCCTGCTCCTGGCCTGCATCGCCGTGGACCGCTACCTGGCCATTGTCCACGCCGTCCATGCCTACCGCCACCGCCGCCTCCTC TCCATCCACATCACCTGTGGGACCATCTGGCTGGTGGGCTTCCTCCTTGCCTTGCCAGAGATTCTCTTCGCCAAAGTCAGCCAAGGCCAT CACAACAACTCCCTGCCACGTTGCACCTTCTCCCAAGAGAACCAAGCAGAAACGCATGCCTGGTTCACCTCCCGATTCCTCTACCATGTG GCGGGATTCCTGCTGCCCATGCTGGTGATGGGCTGGTGCTACGTGGGGGTAGTGCACAGGTTGCGCCAGGCCCAGCGGCGCCCTCAGCGG CAGAAGGCAGTCAGGGTGGCCATCCTGGTGACAAGCATCTTCTTCCTCTGCTGGTCACCCTACCACATCGTCATCTTCCTGGACACCCTG GCGAGGCTGAAGGCCGTGGACAATACCTGCAAGCTGAATGGCTCTCTCCCCGTGGCCATCACCATGTGTGAGTTCCTGGGCCTGGCCCAC TGCTGCCTCAACCCCATGCTCTACACTTTCGCCGGCGTGAAGTTCCGCAGTGACCTGTCGCGGCTCCTGACGAAGCTGGGCTGTACCGGC CCTGCCTCCCTGTGCCAGCTCTTCCCTAGCTGGCGCAGGAGCAGTCTCTCTGAGTCAGAGAATGCCACCTCTCTCACCACGTTCTAGGTC CCAGTGTCCCCTTTTATTGCTGCTTTTCCTTGGGGCAGGCAGTGATGCTGGATGCTCCTTCCAACAGGAGCTGGGATCCTAAGGGCTCAC CGTGGCTAAGAGTGTCCTAGGAGTATCCTCATTTGGGGTAGCTAGAGGAACCAACCCCCATTTCTAGAACATCCCTGCCAGCTCTTCTGC CGGCCCTGGGGCTAGGCTGGAGCCCAGGGAGCGGAAAGCAGCTCAAAGGCACAGTGAAGGCTGTCCTTACCCATCTGCACCCCCCTGGGC TGAGAGAACCTCACGCACCTCCCATCCTAATCATCCAATGCTCAAGAAACAACTTCTACTTCTGCCCTTGCCAACGGAGAGCGCCTGCCC CTCCCAGAACACACTCCATCAGCTTAGGGGCTGCTGACCTCCACAGCTTCCCCTCTCTCCTCCTGCCCACCTGTCAAACAAAGCCAGAAG CTGAGCACCAGGGGATGAGTGGAGGTTAAGGCTGAGGAAAGGCCAGCTGGCAGCAGAGTGTGGCCTTCGGACAACTCAGTCCCTAAAAAC ACAGACATTCTGCCAGGCCCCCAAGCCTGCAGTCATCTTGACCAAGCAGGAAGCTCAGACTGGTTGAGTTCAGGTAGCTGCCCCTGGCTC TGACCGAAACAGCGCTGGGTCCACCCCATGTCACCGGATCCTGGGTGGTCTGCAGGCAGGGCTGACTCTAGGTGCCCTTGGAGGCCAGCC AGTGACCTGAGGAAGCGTGAAGGCCGAGAAGCAAGAAAGAAACCCGACAGAGGGAAGAAAAGAGCTTTCTTCCCGAACCCCAAGGAGGGA GATGGATCAATCAAACCCGGCGGTCCCCTCCGCCAGGCGAGATGGGGTGGGGTGGAGAACTCCTAGGGTGGCTGGGTCCAGGGGATGGGA GGTTGTGGGCATTGATGGGGAAGGAGGCTGGCTTGTCCCCTCCTCACTCCCTTCCCATAAGCTATAGACCCGAGGAAACTCAGAGTCGGA ACGGAGAAAGGTGGACTGGAAGGGGCCCGTGGGAGTCATCTCAACCATCCCCTCCGTGGCATCACCTTAGGCAGGGAAGTGTAAGAAACA CACTGAGGCAGGGAAGTCCCCAGGCCCCAGGAAGCCGTGCCCTGCCCCCGTGAGGATGTCACTCAGATGGAACCGCAGGAAGCTGCTCCG TGCTTGTTTGCTCACCTGGGGTGTGGGAGGCCCGTCCGGCAGTTCTGGGTGCTCCCTACCACCTCCCCAGCCTTTGATCAGGTGGGGAGT CAGGGACCCCTGCCCTTGTCCCACTCAAGCCAAGCAGCCAAGCTCCTTGGGAGGCCCCACTGGGGAAATAACAGCTGTGGCTCACGTGAG AGTGTCTTCACGGCAGGACAACGAGGAAGCCCTAAGACGTCCCTTTTTTCTCTGAGTATCTCCTCGCAAGCTGGGTAATCGATGGGGGAG TCTGAAGCAGATGCAAAGAGGCAAGAGGCTGGATTTTGAATTTTCTTTTTAATAAAAAGGCACCTATAAAACAGGTCAATACAGTACAGG CAGCACAGAGACCCCCGGAACAAGCCTAAAAATTGTTTCAAAATAAAAACCAAGAAGATGTCTTCACATATTGTATTTATATATTTATAT TTATATATATATTTATATAATGGTACAAAATGGCTGGGGGTGTGGCCATGGATGGAGGGAAGAGTAGGCTGGCCTGTGGCGTGGGTGGGA GGAGAGGGGACGGAGAGGGCACTCGGCCCGCTGCAATCTGACCCCTCTCTCCTCAGGGCAGGAAACACAGAGTCAGACAGTTTGGGGGGG TCTTGGGCCAGGGGTGGAGGGCTCAAGGGCACAGGGCCCAGGCTGAGGCAGGGCGGGCAAAGCGCCTGGCAGGATGAAGGGCAAGTGGCC CCCCAAACACAGAGGCCCTGGCCATGGACCCTGGGAGGTGACCGGGGTGAGTCAGGGGCCTGTTGTCAGCCCCAGAGGAAGCGCTGGACC TGGCCGATGGTGGGCCGAGAGGACAGCACCAGGCTGGGAGAAGTGGGGCGAGTTCCCTTTGTATTACAGCTGCCAGTGCAAGACCAGGCC CTCCAGGCCAGGAAGGCTAGGGACGGGTCCTGGTAGAAGACACCCTGTCTAGAATGGCCCTTGGTCCTGGAGGTGGGGCGCAAAAGGCCT CAGCCAGGGAACTGCCCTGCCACCTCCCGAGGCAGGAAAGGAAGTGAGAAAAGGAGAAGTTTTTTTACTCCTGGGGCCAAAGTAGGGGGA CAAACACCCAGTCGTATATGGCTTCAGCTCTGACCAAAGGCGGATAGGGAGCTCTCCTGGGTAGGAGCAGGGAGCCAAGGGGGAGGCAGT GGCTGTGCCTGGGTGGGCACAGACAGATCTGGTACATGGCCCTGAGCCCTGGGCAGAGGGACAACCTTGCCGGTGAGTGGGCAGGCAGAG AGGAGGCGGCAGGATGCTGTTTCCCCGATTCCATCCTCAGGGAGTGGAGACTGGAGGGGAGGTGCACTGACTCAGATGAACTGTTCTCCC CCTTCTTTGATAAGAAGTAGGTGGCAGCAGCCTCTGGAAAAGTCAGGGCCCTGGAGGTTACCTGGCCCAGGGCTACTACAGCCACAGGCC CCAGTGGCACCATGCCACCCCTTCCATGGCTCCACTCAAGGGGGCCACACAGCCACCGCCTCTTCCTCCTTTCCTTCATCCCAAACTGGG ACAAAAGACTTCAAGTTCTGGCTAAGATGTAGCAGCAGCGGATGCCCGGGCATCCAAAGTGGAAAGCCAGGGCCCCGTGTCACCGGTGTG GGCAAACACACATGCACGTGCACACATGTTCTCCCTGAATCACTCAGCAGCAGACAGGCTGCCGCCCTGGGGGTCTCAGCCCTGCTAGGG CTCACCAGGTGGAAGCCTAGGTGGTCTGACCTCAGTTTAGGAGTGGGTCATTTACGTCATCTTACCATTTGGGGACGAGACAGGAATGGT ATCCCTTAGGGACCCAGAGACACTGCAAACAGTGGGTGGCCATGTAGGGCTGCATGTCCCTGGGTCCAGGGGAATGGAGGGAGCAATAAC TTGAAGAAGGGGGGAAGGGTTTCTTTTATCCTTTTTTTTTTGTGTGACTTCTATCAAAACACAGAAATACAGC >13396_13396_1_CBL-CXCR5_CBL_chr11_119103405_ENST00000264033_CXCR5_chr11_118764305_ENST00000292174_length(amino acids)=352AA_BP= MDRLDNYNDTSLVENHLCPATEGPLMASFKAVFVPVAYSLIFLLGVIGNVLVLVILERHRQTRSSTETFLFHLAVADLLLVFILPFAVAE GSVGWVLGTFLCKTVIALHKVNFYCSSLLLACIAVDRYLAIVHAVHAYRHRRLLSIHITCGTIWLVGFLLALPEILFAKVSQGHHNNSLP RCTFSQENQAETHAWFTSRFLYHVAGFLLPMLVMGWCYVGVVHRLRQAQRRPQRQKAVRVAILVTSIFFLCWSPYHIVIFLDTLARLKAV DNTCKLNGSLPVAITMCEFLGLAHCCLNPMLYTFAGVKFRSDLSRLLTKLGCTGPASLCQLFPSWRRSSLSESENATSLTTF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CBL-CXCR5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | CBL | chr11:119103405 | chr11:118764305 | ENST00000264033 | + | 2 | 16 | 648_906 | 147.66666666666666 | 907.0 | CD2AP |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CBL-CXCR5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CBL-CXCR5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CBL | C3150803 | NOONAN SYNDROME-LIKE DISORDER WITH OR WITHOUT JUVENILE MYELOMONOCYTIC LEUKEMIA | 8 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | CBL | C0349639 | Juvenile Myelomonocytic Leukemia | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | CBL | C0008073 | Developmental Disabilities | 1 | CTD_human |
| Hgene | CBL | C0010417 | Cryptorchidism | 1 | CTD_human |
| Hgene | CBL | C0018273 | Growth Disorders | 1 | CTD_human |
| Hgene | CBL | C0021364 | Male infertility | 1 | CTD_human |
| Hgene | CBL | C0028326 | Noonan Syndrome | 1 | GENOMICS_ENGLAND |
| Hgene | CBL | C0042384 | Vasculitis | 1 | CTD_human |
| Hgene | CBL | C0085996 | Child Development Deviations | 1 | CTD_human |
| Hgene | CBL | C0085997 | Child Development Disorders, Specific | 1 | CTD_human |
| Hgene | CBL | C0431663 | Bilateral Cryptorchidism | 1 | CTD_human |
| Hgene | CBL | C0431664 | Unilateral Cryptorchidism | 1 | CTD_human |
| Hgene | CBL | C0848676 | Subfertility, Male | 1 | CTD_human |
| Hgene | CBL | C0917731 | Male sterility | 1 | CTD_human |
| Hgene | CBL | C1563730 | Abdominal Cryptorchidism | 1 | CTD_human |
| Hgene | CBL | C1563731 | Inguinal Cryptorchidism | 1 | CTD_human |
| Hgene | CBL | C4230920 | Fetal hydrops (in some patients) | 1 | GENOMICS_ENGLAND |
| Tgene | C0008312 | Primary biliary cirrhosis | 1 | CTD_human | |
| Tgene | C0023892 | Biliary cirrhosis | 1 | CTD_human | |
| Tgene | C0238065 | Secondary Biliary Cholangitis | 1 | CTD_human | |
| Tgene | C4551595 | Biliary Cirrhosis, Primary, 1 | 1 | CTD_human |
