
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:GTF2IRD1-ALK (FusionGDB2 ID:HG9569TG238) |
Fusion Gene Summary for GTF2IRD1-ALK |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: GTF2IRD1-ALK | Fusion gene ID: hg9569tg238 | Hgene | Tgene | Gene symbol | GTF2IRD1 | ALK | Gene ID | 9569 | 238 |
| Gene name | GTF2I repeat domain containing 1 | ALK receptor tyrosine kinase | |
| Synonyms | BEN|CREAM1|GTF3|MUSTRD1|RBAP2|WBS|WBSCR11|WBSCR12|hMusTRD1alpha1 | CD246|NBLST3 | |
| Cytomap | ('GTF2IRD1')('ALK') 7q11.23 | 2p23.2-p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | general transcription factor II-I repeat domain-containing protein 1USE B1-binding proteinWilliams-Beuren syndrome chromosome region 11binding factor for early enhancergeneral transcription factor 3general transcription factor IIImuscle TFII-I repea | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | Q9UM73 | |
| Ensembl transtripts involved in fusion gene | ENST00000489094, ENST00000265755, ENST00000424337, ENST00000455841, ENST00000476977, | ||
| Fusion gene scores | * DoF score | 11 X 6 X 9=594 | 56 X 74 X 20=82880 |
| # samples | 12 | 57 | |
| ** MAII score | log2(12/594*10)=-2.30742852519225 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(57/82880*10)=-7.18391827352181 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: GTF2IRD1 [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | GTF2IRD1(73935627)-ALK(29446394), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | GTF2IRD1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GTF2IRD1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GTF2IRD1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. GTF2IRD1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Fusion gene breakpoints across GTF2IRD1 (5'-gene) Fusion gene breakpoints across GTF2IRD1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
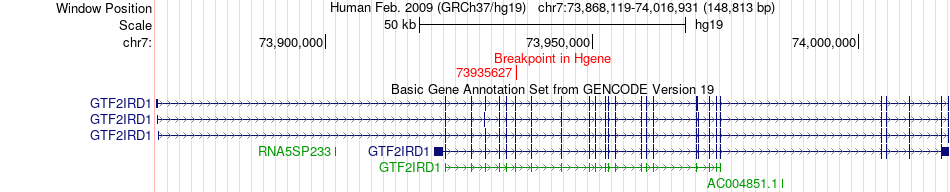 |
 Fusion gene breakpoints across ALK (3'-gene) Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
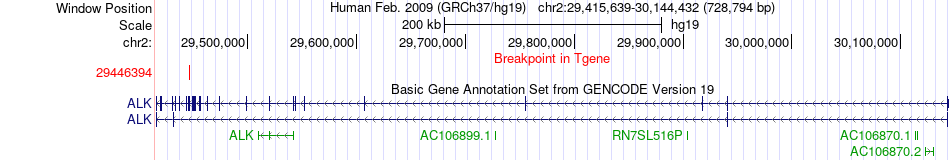 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-EL-A4KD-01A | GTF2IRD1 | chr7 | 73935627 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | THCA | TCGA-EL-A4KD-01A | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
Top |
Fusion Gene ORF analysis for GTF2IRD1-ALK |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000489094 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 3UTR-intron | ENST00000489094 | ENST00000431873 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 3UTR-intron | ENST00000489094 | ENST00000498037 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000265755 | ENST00000431873 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000265755 | ENST00000498037 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000424337 | ENST00000431873 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000424337 | ENST00000498037 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000455841 | ENST00000431873 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000455841 | ENST00000498037 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000476977 | ENST00000431873 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| 5CDS-intron | ENST00000476977 | ENST00000498037 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| In-frame | ENST00000265755 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| In-frame | ENST00000424337 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| In-frame | ENST00000455841 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
| In-frame | ENST00000476977 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000265755 | GTF2IRD1 | chr7 | 73935627 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3540 | 1399 | 330 | 3089 | 919 |
| ENST00000455841 | GTF2IRD1 | chr7 | 73935627 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3456 | 1315 | 150 | 3005 | 951 |
| ENST00000424337 | GTF2IRD1 | chr7 | 73935627 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3221 | 1080 | 11 | 2770 | 919 |
| ENST00000476977 | GTF2IRD1 | chr7 | 73935627 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4838 | 2697 | 1691 | 4387 | 898 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000265755 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - | 0.005352864 | 0.9946471 |
| ENST00000455841 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - | 0.004399282 | 0.9956007 |
| ENST00000424337 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - | 0.005383978 | 0.994616 |
| ENST00000476977 | ENST00000389048 | GTF2IRD1 | chr7 | 73935627 | + | ALK | chr2 | 29446394 | - | 0.01181 | 0.98819005 |
Top |
Fusion Genomic Features for GTF2IRD1-ALK |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
Top |
Fusion Protein Features for GTF2IRD1-ALK |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:73935627/chr2:29446394) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ALK |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Neuronal receptor tyrosine kinase that is essentially and transiently expressed in specific regions of the central and peripheral nervous systems and plays an important role in the genesis and differentiation of the nervous system. Transduces signals from ligands at the cell surface, through specific activation of the mitogen-activated protein kinase (MAPK) pathway. Phosphorylates almost exclusively at the first tyrosine of the Y-x-x-x-Y-Y motif. Following activation by ligand, ALK induces tyrosine phosphorylation of CBL, FRS2, IRS1 and SHC1, as well as of the MAP kinases MAPK1/ERK2 and MAPK3/ERK1. Acts as a receptor for ligands pleiotrophin (PTN), a secreted growth factor, and midkine (MDK), a PTN-related factor, thus participating in PTN and MDK signal transduction. PTN-binding induces MAPK pathway activation, which is important for the anti-apoptotic signaling of PTN and regulation of cell proliferation. MDK-binding induces phosphorylation of the ALK target insulin receptor substrate (IRS1), activates mitogen-activated protein kinases (MAPKs) and PI3-kinase, resulting also in cell proliferation induction. Drives NF-kappa-B activation, probably through IRS1 and the activation of the AKT serine/threonine kinase. Recruitment of IRS1 to activated ALK and the activation of NF-kappa-B are essential for the autocrine growth and survival signaling of MDK. Thinness gene involved in the resistance to weight gain: in hypothalamic neurons, controls energy expenditure acting as a negative regulator of white adipose tissue lipolysis and sympathetic tone to fine-tune energy homeostasis (By similarity). {ECO:0000250|UniProtKB:P97793, ECO:0000269|PubMed:11121404, ECO:0000269|PubMed:11278720, ECO:0000269|PubMed:11387242, ECO:0000269|PubMed:11809760, ECO:0000269|PubMed:12107166, ECO:0000269|PubMed:12122009, ECO:0000269|PubMed:15226403, ECO:0000269|PubMed:15908427, ECO:0000269|PubMed:16317043, ECO:0000269|PubMed:16878150, ECO:0000269|PubMed:17274988}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 119_213 | 335 | 960.0 | Repeat | Note=GTF2I-like 1 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 119_213 | 335 | 945.0 | Repeat | Note=GTF2I-like 1 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 119_213 | 367 | 977.0 | Repeat | Note=GTF2I-like 1 |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057 | 1621.0 | Topological domain | Cytoplasmic |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 906_930 | 335 | 960.0 | Compositional bias | Note=Ser-rich |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 906_930 | 335 | 945.0 | Compositional bias | Note=Ser-rich |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 906_930 | 367 | 977.0 | Compositional bias | Note=Ser-rich |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 898_905 | 335 | 960.0 | Motif | Nuclear localization signal |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 898_905 | 335 | 945.0 | Motif | Nuclear localization signal |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 898_905 | 367 | 977.0 | Motif | Nuclear localization signal |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 342_436 | 335 | 960.0 | Repeat | Note=GTF2I-like 2 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 556_650 | 335 | 960.0 | Repeat | Note=GTF2I-like 3 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 696_790 | 335 | 960.0 | Repeat | Note=GTF2I-like 4 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000265755 | + | 7 | 27 | 793_887 | 335 | 960.0 | Repeat | Note=GTF2I-like 5 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 342_436 | 335 | 945.0 | Repeat | Note=GTF2I-like 2 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 556_650 | 335 | 945.0 | Repeat | Note=GTF2I-like 3 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 696_790 | 335 | 945.0 | Repeat | Note=GTF2I-like 4 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000424337 | + | 7 | 27 | 793_887 | 335 | 945.0 | Repeat | Note=GTF2I-like 5 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 342_436 | 367 | 977.0 | Repeat | Note=GTF2I-like 2 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 556_650 | 367 | 977.0 | Repeat | Note=GTF2I-like 3 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 696_790 | 367 | 977.0 | Repeat | Note=GTF2I-like 4 |
| Hgene | GTF2IRD1 | chr7:73935627 | chr2:29446394 | ENST00000455841 | + | 7 | 27 | 793_887 | 367 | 977.0 | Repeat | Note=GTF2I-like 5 |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr7:73935627 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057 | 1621.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for GTF2IRD1-ALK |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >35245_35245_1_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000265755_ALK_chr2_29446394_ENST00000389048_length(transcript)=3540nt_BP=1399nt TAAATGGCAGCCAATGGAGGGTGGTGTTGCGCGGGGCTGGGATTAGGGCCGGGGCGAATGGCTGGCAATCTTACTGGGATTACAGAACAA AGAGCCTCCCCGCGCTCCCGCTCTCCGCTCCTCTCCCCGCGCCGCCCCGCCCTCCGCCGCAGCCCGCGCCGGGGGTGGGGGCCGCCGAGC GCCAGCCCCCCGGCCGGCCGATTCCCCCCCCGCGCCCCCTCCCCGCGCCTCCCTCCCCGCCCTCGCCGCGCCGCCGTCCTCGCCTCCCTC TGCCTCTCCTTCCCCCATTCTCCCGGATTAATTAAGGAGGCAGCGGCAGGAGGCTGAGTCCTGGCCGCGGGCCGGGGCCGGGGCGCCGCT GGCAGGAGCGCTTGGGGATCCTCCAAGGCGACCATGGCCTTGCTGGGTAAGCGCTGTGACGTCCCCACCAACGGCTGCGGACCCGACCGC TGGAACTCCGCGTTCACCCGCAAAGACGAGATCATCACCAGCCTCGTGTCTGCCTTAGACTCCATGTGCTCAGCGCTGTCCAAACTGAAC GCCGAGGTGGCCTGTGTCGCCGTGCACGATGAGAGCGCCTTTGTGGTGGGCACAGAGAAGGGGAGAATGTTCCTGAATGCCCGGAAGGAG CTACAGTCAGACTTCCTCAGGTTCTGCCGAGGGCCCCCGTGGAAGGATCCGGAGGCAGAGCACCCCAAGAAGGTGCAGCGGGGCGAGGGT GGAGGCCGTAGCCTCCCTCGGTCCTCCCTGGAACATGGCTCAGATGTGTACCTTCTGCGGAAGATGGTAGAGGAGGTGTTTGATGTTCTT TATAGCGAGGCCCTGGGAAGGGCCAGTGTGGTGCCACTGCCCTATGAGAGGCTGCTCAGGGAGCCAGGGCTGCTGGCCGTGCAGGGGCTG CCCGAAGGCCTGGCCTTCCGAAGGCCAGCCGAGTATGACCCCAAGGCCCTCATGGCCATCCTGGAACACAGCCACCGCATCCGCTTCAAG CTCAAGAGGCCACTTGAGGATGGCGGGCGGGACTCGAAGGCCCTGGTGGAGCTGAACGGTGTCTCCCTGATTCCCAAGGGGTCACGGGAC TGTGGCCTGCATGGCCAGGCCCCCAAGGTGCCACCCCAGGACCTGCCCCCAACCGCCACCTCCTCCTCCATGGCCAGCTTCCTGTACAGC ACGGCGCTCCCCAACCACGCCATCCGAGAGCTCAAGCAGGAAGCACCTTCCTGCCCCCTTGCCCCCAGCGACCTGGGCCTGAGTCGGCCC ATGCCAGAGCCCAAGGCCACCGGTGCCCAAGACTTCTCCGACTGTTGTGGACAGAAGCCCACTGGGCCTGGTGGGCCTCTCATCCAGAAC GTCCATGCCTCCAAGCGCATTCTCTTCTCCATCGTCCATGACAAGTCAGTGTACCGCCGGAAGCACCAGGAGCTGCAAGCCATGCAGATG GAGCTGCAGAGCCCTGAGTACAAGCTGAGCAAGCTCCGCACCTCGACCATCATGACCGACTACAACCCCAACTACTGCTTTGCTGGCAAG ACCTCCTCCATCAGTGACCTGAAGGAGGTGCCGCGGAAAAACATCACCCTCATTCGGGGTCTGGGCCATGGCGCCTTTGGGGAGGTGTAT GAAGGCCAGGTGTCCGGAATGCCCAACGACCCAAGCCCCCTGCAAGTGGCTGTGAAGACGCTGCCTGAAGTGTGCTCTGAACAGGACGAA CTGGATTTCCTCATGGAAGCCCTGATCATCAGCAAATTCAACCACCAGAACATTGTTCGCTGCATTGGGGTGAGCCTGCAATCCCTGCCC CGGTTCATCCTGCTGGAGCTCATGGCGGGGGGAGACCTCAAGTCCTTCCTCCGAGAGACCCGCCCTCGCCCGAGCCAGCCCTCCTCCCTG GCCATGCTGGACCTTCTGCACGTGGCTCGGGACATTGCCTGTGGCTGTCAGTATTTGGAGGAAAACCACTTCATCCACCGAGACATTGCT GCCAGAAACTGCCTCTTGACCTGTCCAGGCCCTGGAAGAGTGGCCAAGATTGGAGACTTCGGGATGGCCCGAGACATCTACAGGGCGAGC TACTATAGAAAGGGAGGCTGTGCCATGCTGCCAGTTAAGTGGATGCCCCCAGAGGCCTTCATGGAAGGAATATTCACTTCTAAAACAGAC ACATGGTCCTTTGGAGTGCTGCTATGGGAAATCTTTTCTCTTGGATATATGCCATACCCCAGCAAAAGCAACCAGGAAGTTCTGGAGTTT GTCACCAGTGGAGGCCGGATGGACCCACCCAAGAACTGCCCTGGGCCTGTATACCGGATAATGACTCAGTGCTGGCAACATCAGCCTGAA GACAGGCCCAACTTTGCCATCATTTTGGAGAGGATTGAATACTGCACCCAGGACCCGGATGTAATCAACACCGCTTTGCCGATAGAATAT GGTCCACTTGTGGAAGAGGAAGAGAAAGTGCCTGTGAGGCCCAAGGACCCTGAGGGGGTTCCTCCTCTCCTGGTCTCTCAACAGGCAAAA CGGGAGGAGGAGCGCAGCCCAGCTGCCCCACCACCTCTGCCTACCACCTCCTCTGGCAAGGCTGCAAAGAAACCCACAGCTGCAGAGATC TCTGTTCGAGTCCCTAGAGGGCCGGCCGTGGAAGGGGGACACGTGAATATGGCATTCTCTCAGTCCAACCCTCCTTCGGAGTTGCACAAG GTCCACGGATCCAGAAACAAGCCCACCAGCTTGTGGAACCCAACGTACGGCTCCTGGTTTACAGAGAAACCCACCAAAAAGAATAATCCT ATAGCAAAGAAGGAGCCACACGACAGGGGTAACCTGGGGCTGGAGGGAAGCTGTACTGTCCCACCTAACGTTGCAACTGGGAGACTTCCG GGGGCCTCACTGCTCCTAGAGCCCTCTTCGCTGACTGCCAATATGAAGGAGGTACCTCTGTTCAGGCTACGTCACTTCCCTTGTGGGAAT GTCAATTACGGCTACCAGCAACAGGGCTTGCCCTTAGAAGCCGCTACTGCCCCTGGAGCTGGTCATTACGAGGATACCATTCTGAAAAGC AAGAATAGCATGAACCAGCCTGGGCCCTGAGCTCGGTCGCACACTCACTTCTCTTCCTTGGGATCCCTAAGACCGTGGAGGAGAGAGAGG CAATGGCTCCTTCACAAACCAGAGACCAAATGTCACGTTTTGTTTTGTGCCAACCTATTTTGAAGTACCACCAAAAAAGCTGTATTTTGA AAATGCTTTAGAAAGGTTTTGAGCATGGGTTCATCCTATTCTTTCGAAAGAAGAAAATATCATAAAAATGAGTGATAAATACAAGGCCCA GATGTGGTTGCATAAGGTTTTTATGCATGTTTGTTGTATACTTCCTTATGCTTCTTTCAAATTGTGTGTGCTCTGCTTCAATGTAGTCAG AATTAGCTGCTTCTATGTTTCATAGTTGGGGTCATAGATGTTTCCTTGCCTTGTTGATGTGGACATGAGCCATTTGAGGGGAGAGGGAAC GGAAATAAAGGAGTTATTTGTAATGACTAA >35245_35245_1_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000265755_ALK_chr2_29446394_ENST00000389048_length(amino acids)=919AA_BP=356 MAAGRGRGAAGRSAWGSSKATMALLGKRCDVPTNGCGPDRWNSAFTRKDEIITSLVSALDSMCSALSKLNAEVACVAVHDESAFVVGTEK GRMFLNARKELQSDFLRFCRGPPWKDPEAEHPKKVQRGEGGGRSLPRSSLEHGSDVYLLRKMVEEVFDVLYSEALGRASVVPLPYERLLR EPGLLAVQGLPEGLAFRRPAEYDPKALMAILEHSHRIRFKLKRPLEDGGRDSKALVELNGVSLIPKGSRDCGLHGQAPKVPPQDLPPTAT SSSMASFLYSTALPNHAIRELKQEAPSCPLAPSDLGLSRPMPEPKATGAQDFSDCCGQKPTGPGGPLIQNVHASKRILFSIVHDKSVYRR KHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKT LPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLE ENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYP SKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGV PPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWF TEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGA GHYEDTILKSKNSMNQPGP -------------------------------------------------------------- >35245_35245_2_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000424337_ALK_chr2_29446394_ENST00000389048_length(transcript)=3221nt_BP=1080nt GAGGCTGAGTCCTGGCCGCGGGCCGGGGCCGGGGCGCCGCTGGCAGGAGCGCTTGGGGATCCTCCAAGGCGACCATGGCCTTGCTGGGTA AGCGCTGTGACGTCCCCACCAACGGCTGCGGACCCGACCGCTGGAACTCCGCGTTCACCCGCAAAGACGAGATCATCACCAGCCTCGTGT CTGCCTTAGACTCCATGTGCTCAGCGCTGTCCAAACTGAACGCCGAGGTGGCCTGTGTCGCCGTGCACGATGAGAGCGCCTTTGTGGTGG GCACAGAGAAGGGGAGAATGTTCCTGAATGCCCGGAAGGAGCTACAGTCAGACTTCCTCAGGTTCTGCCGAGGGCCCCCGTGGAAGGATC CGGAGGCAGAGCACCCCAAGAAGGTGCAGCGGGGCGAGGGTGGAGGCCGTAGCCTCCCTCGGTCCTCCCTGGAACATGGCTCAGATGTGT ACCTTCTGCGGAAGATGGTAGAGGAGGTGTTTGATGTTCTTTATAGCGAGGCCCTGGGAAGGGCCAGTGTGGTGCCACTGCCCTATGAGA GGCTGCTCAGGGAGCCAGGGCTGCTGGCCGTGCAGGGGCTGCCCGAAGGCCTGGCCTTCCGAAGGCCAGCCGAGTATGACCCCAAGGCCC TCATGGCCATCCTGGAACACAGCCACCGCATCCGCTTCAAGCTCAAGAGGCCACTTGAGGATGGCGGGCGGGACTCGAAGGCCCTGGTGG AGCTGAACGGTGTCTCCCTGATTCCCAAGGGGTCACGGGACTGTGGCCTGCATGGCCAGGCCCCCAAGGTGCCACCCCAGGACCTGCCCC CAACCGCCACCTCCTCCTCCATGGCCAGCTTCCTGTACAGCACGGCGCTCCCCAACCACGCCATCCGAGAGCTCAAGCAGGAAGCACCTT CCTGCCCCCTTGCCCCCAGCGACCTGGGCCTGAGTCGGCCCATGCCAGAGCCCAAGGCCACCGGTGCCCAAGACTTCTCCGACTGTTGTG GACAGAAGCCCACTGGGCCTGGTGGGCCTCTCATCCAGAACGTCCATGCCTCCAAGCGCATTCTCTTCTCCATCGTCCATGACAAGTCAG TGTACCGCCGGAAGCACCAGGAGCTGCAAGCCATGCAGATGGAGCTGCAGAGCCCTGAGTACAAGCTGAGCAAGCTCCGCACCTCGACCA TCATGACCGACTACAACCCCAACTACTGCTTTGCTGGCAAGACCTCCTCCATCAGTGACCTGAAGGAGGTGCCGCGGAAAAACATCACCC TCATTCGGGGTCTGGGCCATGGCGCCTTTGGGGAGGTGTATGAAGGCCAGGTGTCCGGAATGCCCAACGACCCAAGCCCCCTGCAAGTGG CTGTGAAGACGCTGCCTGAAGTGTGCTCTGAACAGGACGAACTGGATTTCCTCATGGAAGCCCTGATCATCAGCAAATTCAACCACCAGA ACATTGTTCGCTGCATTGGGGTGAGCCTGCAATCCCTGCCCCGGTTCATCCTGCTGGAGCTCATGGCGGGGGGAGACCTCAAGTCCTTCC TCCGAGAGACCCGCCCTCGCCCGAGCCAGCCCTCCTCCCTGGCCATGCTGGACCTTCTGCACGTGGCTCGGGACATTGCCTGTGGCTGTC AGTATTTGGAGGAAAACCACTTCATCCACCGAGACATTGCTGCCAGAAACTGCCTCTTGACCTGTCCAGGCCCTGGAAGAGTGGCCAAGA TTGGAGACTTCGGGATGGCCCGAGACATCTACAGGGCGAGCTACTATAGAAAGGGAGGCTGTGCCATGCTGCCAGTTAAGTGGATGCCCC CAGAGGCCTTCATGGAAGGAATATTCACTTCTAAAACAGACACATGGTCCTTTGGAGTGCTGCTATGGGAAATCTTTTCTCTTGGATATA TGCCATACCCCAGCAAAAGCAACCAGGAAGTTCTGGAGTTTGTCACCAGTGGAGGCCGGATGGACCCACCCAAGAACTGCCCTGGGCCTG TATACCGGATAATGACTCAGTGCTGGCAACATCAGCCTGAAGACAGGCCCAACTTTGCCATCATTTTGGAGAGGATTGAATACTGCACCC AGGACCCGGATGTAATCAACACCGCTTTGCCGATAGAATATGGTCCACTTGTGGAAGAGGAAGAGAAAGTGCCTGTGAGGCCCAAGGACC CTGAGGGGGTTCCTCCTCTCCTGGTCTCTCAACAGGCAAAACGGGAGGAGGAGCGCAGCCCAGCTGCCCCACCACCTCTGCCTACCACCT CCTCTGGCAAGGCTGCAAAGAAACCCACAGCTGCAGAGATCTCTGTTCGAGTCCCTAGAGGGCCGGCCGTGGAAGGGGGACACGTGAATA TGGCATTCTCTCAGTCCAACCCTCCTTCGGAGTTGCACAAGGTCCACGGATCCAGAAACAAGCCCACCAGCTTGTGGAACCCAACGTACG GCTCCTGGTTTACAGAGAAACCCACCAAAAAGAATAATCCTATAGCAAAGAAGGAGCCACACGACAGGGGTAACCTGGGGCTGGAGGGAA GCTGTACTGTCCCACCTAACGTTGCAACTGGGAGACTTCCGGGGGCCTCACTGCTCCTAGAGCCCTCTTCGCTGACTGCCAATATGAAGG AGGTACCTCTGTTCAGGCTACGTCACTTCCCTTGTGGGAATGTCAATTACGGCTACCAGCAACAGGGCTTGCCCTTAGAAGCCGCTACTG CCCCTGGAGCTGGTCATTACGAGGATACCATTCTGAAAAGCAAGAATAGCATGAACCAGCCTGGGCCCTGAGCTCGGTCGCACACTCACT TCTCTTCCTTGGGATCCCTAAGACCGTGGAGGAGAGAGAGGCAATGGCTCCTTCACAAACCAGAGACCAAATGTCACGTTTTGTTTTGTG CCAACCTATTTTGAAGTACCACCAAAAAAGCTGTATTTTGAAAATGCTTTAGAAAGGTTTTGAGCATGGGTTCATCCTATTCTTTCGAAA GAAGAAAATATCATAAAAATGAGTGATAAATACAAGGCCCAGATGTGGTTGCATAAGGTTTTTATGCATGTTTGTTGTATACTTCCTTAT GCTTCTTTCAAATTGTGTGTGCTCTGCTTCAATGTAGTCAGAATTAGCTGCTTCTATGTTTCATAGTTGGGGTCATAGATGTTTCCTTGC CTTGTTGATGTGGACATGAGCCATTTGAGGGGAGAGGGAACGGAAATAAAGGAGTTATTTGTAATGACTAA >35245_35245_2_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000424337_ALK_chr2_29446394_ENST00000389048_length(amino acids)=919AA_BP=356 MAAGRGRGAAGRSAWGSSKATMALLGKRCDVPTNGCGPDRWNSAFTRKDEIITSLVSALDSMCSALSKLNAEVACVAVHDESAFVVGTEK GRMFLNARKELQSDFLRFCRGPPWKDPEAEHPKKVQRGEGGGRSLPRSSLEHGSDVYLLRKMVEEVFDVLYSEALGRASVVPLPYERLLR EPGLLAVQGLPEGLAFRRPAEYDPKALMAILEHSHRIRFKLKRPLEDGGRDSKALVELNGVSLIPKGSRDCGLHGQAPKVPPQDLPPTAT SSSMASFLYSTALPNHAIRELKQEAPSCPLAPSDLGLSRPMPEPKATGAQDFSDCCGQKPTGPGGPLIQNVHASKRILFSIVHDKSVYRR KHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKT LPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLE ENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYP SKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGV PPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWF TEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGA GHYEDTILKSKNSMNQPGP -------------------------------------------------------------- >35245_35245_3_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000455841_ALK_chr2_29446394_ENST00000389048_length(transcript)=3456nt_BP=1315nt GCCAGCCCCCCGGCCGGCCGATTCCCCCCCCGCGCCCCCTCCCCGCGCCTCCCTCCCCGCCCTCGCCGCGCCGCCGTCCTCGCCTCCCTC TGCCTCTCCTTCCCCCATTCTCCCGGATTAATTAAGGAGGCAGCGGCAGGAGGCTGAGTCCTGGCCGCGGGCCGGGGCCGGGGCGCCGCT GGCAGGAGCGCTTGGGGATCCTCCAAGGCGACCATGGCCTTGCTGGGTAAGCGCTGTGACGTCCCCACCAACGGCTGCGGACCCGACCGC TGGAACTCCGCGTTCACCCGCAAAGACGAGATCATCACCAGCCTCGTGTCTGCCTTAGACTCCATGTGCTCAGCGCTGTCCAAACTGAAC GCCGAGGTGGCCTGTGTCGCCGTGCACGATGAGAGCGCCTTTGTGGTGGGCACAGAGAAGGGGAGAATGTTCCTGAATGCCCGGAAGGAG CTACAGTCAGACTTCCTCAGGTTCTGCCTCTCCGCAGCTCAGCACAGGGCAGCGACATCCCAGCTCGAAGGCCGGGTGGTGAGACGGGTG CTCACTGTGGCCTCGCGTGCTCTGTGTCCCACAGGAGGGCCCCCGTGGAAGGATCCGGAGGCAGAGCACCCCAAGAAGGTGCAGCGGGGC GAGGGTGGAGGCCGTAGCCTCCCTCGGTCCTCCCTGGAACATGGCTCAGATGTGTACCTTCTGCGGAAGATGGTAGAGGAGGTGTTTGAT GTTCTTTATAGCGAGGCCCTGGGAAGGGCCAGTGTGGTGCCACTGCCCTATGAGAGGCTGCTCAGGGAGCCAGGGCTGCTGGCCGTGCAG GGGCTGCCCGAAGGCCTGGCCTTCCGAAGGCCAGCCGAGTATGACCCCAAGGCCCTCATGGCCATCCTGGAACACAGCCACCGCATCCGC TTCAAGCTCAAGAGGCCACTTGAGGATGGCGGGCGGGACTCGAAGGCCCTGGTGGAGCTGAACGGTGTCTCCCTGATTCCCAAGGGGTCA CGGGACTGTGGCCTGCATGGCCAGGCCCCCAAGGTGCCACCCCAGGACCTGCCCCCAACCGCCACCTCCTCCTCCATGGCCAGCTTCCTG TACAGCACGGCGCTCCCCAACCACGCCATCCGAGAGCTCAAGCAGGAAGCACCTTCCTGCCCCCTTGCCCCCAGCGACCTGGGCCTGAGT CGGCCCATGCCAGAGCCCAAGGCCACCGGTGCCCAAGACTTCTCCGACTGTTGTGGACAGAAGCCCACTGGGCCTGGTGGGCCTCTCATC CAGAACGTCCATGCCTCCAAGCGCATTCTCTTCTCCATCGTCCATGACAAGTCAGTGTACCGCCGGAAGCACCAGGAGCTGCAAGCCATG CAGATGGAGCTGCAGAGCCCTGAGTACAAGCTGAGCAAGCTCCGCACCTCGACCATCATGACCGACTACAACCCCAACTACTGCTTTGCT GGCAAGACCTCCTCCATCAGTGACCTGAAGGAGGTGCCGCGGAAAAACATCACCCTCATTCGGGGTCTGGGCCATGGCGCCTTTGGGGAG GTGTATGAAGGCCAGGTGTCCGGAATGCCCAACGACCCAAGCCCCCTGCAAGTGGCTGTGAAGACGCTGCCTGAAGTGTGCTCTGAACAG GACGAACTGGATTTCCTCATGGAAGCCCTGATCATCAGCAAATTCAACCACCAGAACATTGTTCGCTGCATTGGGGTGAGCCTGCAATCC CTGCCCCGGTTCATCCTGCTGGAGCTCATGGCGGGGGGAGACCTCAAGTCCTTCCTCCGAGAGACCCGCCCTCGCCCGAGCCAGCCCTCC TCCCTGGCCATGCTGGACCTTCTGCACGTGGCTCGGGACATTGCCTGTGGCTGTCAGTATTTGGAGGAAAACCACTTCATCCACCGAGAC ATTGCTGCCAGAAACTGCCTCTTGACCTGTCCAGGCCCTGGAAGAGTGGCCAAGATTGGAGACTTCGGGATGGCCCGAGACATCTACAGG GCGAGCTACTATAGAAAGGGAGGCTGTGCCATGCTGCCAGTTAAGTGGATGCCCCCAGAGGCCTTCATGGAAGGAATATTCACTTCTAAA ACAGACACATGGTCCTTTGGAGTGCTGCTATGGGAAATCTTTTCTCTTGGATATATGCCATACCCCAGCAAAAGCAACCAGGAAGTTCTG GAGTTTGTCACCAGTGGAGGCCGGATGGACCCACCCAAGAACTGCCCTGGGCCTGTATACCGGATAATGACTCAGTGCTGGCAACATCAG CCTGAAGACAGGCCCAACTTTGCCATCATTTTGGAGAGGATTGAATACTGCACCCAGGACCCGGATGTAATCAACACCGCTTTGCCGATA GAATATGGTCCACTTGTGGAAGAGGAAGAGAAAGTGCCTGTGAGGCCCAAGGACCCTGAGGGGGTTCCTCCTCTCCTGGTCTCTCAACAG GCAAAACGGGAGGAGGAGCGCAGCCCAGCTGCCCCACCACCTCTGCCTACCACCTCCTCTGGCAAGGCTGCAAAGAAACCCACAGCTGCA GAGATCTCTGTTCGAGTCCCTAGAGGGCCGGCCGTGGAAGGGGGACACGTGAATATGGCATTCTCTCAGTCCAACCCTCCTTCGGAGTTG CACAAGGTCCACGGATCCAGAAACAAGCCCACCAGCTTGTGGAACCCAACGTACGGCTCCTGGTTTACAGAGAAACCCACCAAAAAGAAT AATCCTATAGCAAAGAAGGAGCCACACGACAGGGGTAACCTGGGGCTGGAGGGAAGCTGTACTGTCCCACCTAACGTTGCAACTGGGAGA CTTCCGGGGGCCTCACTGCTCCTAGAGCCCTCTTCGCTGACTGCCAATATGAAGGAGGTACCTCTGTTCAGGCTACGTCACTTCCCTTGT GGGAATGTCAATTACGGCTACCAGCAACAGGGCTTGCCCTTAGAAGCCGCTACTGCCCCTGGAGCTGGTCATTACGAGGATACCATTCTG AAAAGCAAGAATAGCATGAACCAGCCTGGGCCCTGAGCTCGGTCGCACACTCACTTCTCTTCCTTGGGATCCCTAAGACCGTGGAGGAGA GAGAGGCAATGGCTCCTTCACAAACCAGAGACCAAATGTCACGTTTTGTTTTGTGCCAACCTATTTTGAAGTACCACCAAAAAAGCTGTA TTTTGAAAATGCTTTAGAAAGGTTTTGAGCATGGGTTCATCCTATTCTTTCGAAAGAAGAAAATATCATAAAAATGAGTGATAAATACAA GGCCCAGATGTGGTTGCATAAGGTTTTTATGCATGTTTGTTGTATACTTCCTTATGCTTCTTTCAAATTGTGTGTGCTCTGCTTCAATGT AGTCAGAATTAGCTGCTTCTATGTTTCATAGTTGGGGTCATAGATGTTTCCTTGCCTTGTTGATGTGGACATGAGCCATTTGAGGGGAGA GGGAACGGAAATAAAGGAGTTATTTGTAATGACTAA >35245_35245_3_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000455841_ALK_chr2_29446394_ENST00000389048_length(amino acids)=951AA_BP=388 MAAGRGRGAAGRSAWGSSKATMALLGKRCDVPTNGCGPDRWNSAFTRKDEIITSLVSALDSMCSALSKLNAEVACVAVHDESAFVVGTEK GRMFLNARKELQSDFLRFCLSAAQHRAATSQLEGRVVRRVLTVASRALCPTGGPPWKDPEAEHPKKVQRGEGGGRSLPRSSLEHGSDVYL LRKMVEEVFDVLYSEALGRASVVPLPYERLLREPGLLAVQGLPEGLAFRRPAEYDPKALMAILEHSHRIRFKLKRPLEDGGRDSKALVEL NGVSLIPKGSRDCGLHGQAPKVPPQDLPPTATSSSMASFLYSTALPNHAIRELKQEAPSCPLAPSDLGLSRPMPEPKATGAQDFSDCCGQ KPTGPGGPLIQNVHASKRILFSIVHDKSVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLI RGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLR ETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPE AFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQD PDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMA FSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEV PLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQPGP -------------------------------------------------------------- >35245_35245_4_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000476977_ALK_chr2_29446394_ENST00000389048_length(transcript)=4838nt_BP=2697nt AACATTTAGCAGCAAACTCAACATACATTTTGGCCAAACGCCCACCGGCCAGTTGTTTAAATCAATATTTATCCACAGCCAAAAAGCAGA GGAGAGCCAGAACTGAGGCCAGAGCCGAGCTCTGATGCATCTCTCATTTCTCGGGATGTTTCTGTCCCTGTGGTTGGACACCTCTGGCCT TGTGAAGTGTGATGCACTGTCACATCTCCTGTTCTGTGTCATTGGCCAATGATCATATATCATGACCTGCTGGAAGGCCTGTCTGTGGCT GGGACCACACGCCTTGGGCCTTATGCACACTGGGCACTGGTCGGGATCCTGGGGTGCAACAGTGGCAGGCAGACCTGGTTTCTGCCCTCA AAGAGCTTACAGATGGCAGGGGCACCCATGGCGGCAGAAGACACCCCAGGCCTGGTGCCCTTTGTGGTGCCAGCCCCATGGTCCTCCTGC CTGGGCCTTCCCTACCCCATTGGGTGCAGAAACTCCCTGTCTGCAGGTAGGAGACAGAGGGTAGGTTTTCAGGCTCCCTTGGGAACTGCA GCCCTGCTCTCTGCCATCAACACCCAGCAGGGGCCACACAGAGAGCACCGGGACTGAGCCCATAGAGGGGAACCGAGAGGCCCCTGCCTC TAGTCTCTGCCTTCTTTGCTTGGATTGGTGCAGGGACAGCTGCCTTGAGGGCAGGCCCTGGCACTGGGGCAGCCTGTGGGTGCCCCCTGG GTCAAGAAGGAGAGGGGCAGGGTAGAACCAGGAGCCAAAGGAGGCTGATCTTTGCATCTCATGGGTGCCCAGCTGGACACTGTCATACCC AGGAAGCCTGTGCCATGCCATGGGGACCCACAACTGGGGGCCCTGGACTTGAGGGGGAGGATGCAGCTCTGTCCCCCAGGAACCCCATTG CAACAGGACACAGTCCTGCCCTGGGGAGCCCCTGACCTGAGACAAAGCAGCCTCGGCCCTGCTGTATCTTTCCATACCCCTGATGCCAAG TCTCCTGGCTAGGAGGGAAACTGAGGCTGGAAGGCCTCGGCGGGGGTGGCATTGGCCTCGGGAGCATGTGGCTTGATGCAGAAATGTGAC GGCAGAGCTCAGAGGCATGCGGAAGGGAGGGGAGGACATCACCGGCTCCTGACCCAGCTGGGCTTCAGGTTGGGGGTACAGGAGGTGGGC AAGCAGGTTGGACAATTAAAAGCTTCGATGAGGCTGGGTGAGTGGCTTATGCCTGTATTTCCAACACTTTGGGAGGCTGAGGTGGGCAGA TCACCTGAGGCCAGGAGTTCAAGACCAGCCTGGCCAACATGGTGAAACCCCATCTCTACTAAAAATACAAAAATTAGCCAGACGTGTGGT GGCACCTGTAATCCCAGCTACCCGGGAGGCTGAGGCAGGAGAATCACTCGAACCCAGGAAGGGGAGGTTGCAGTGAGCCAAGATTGCACC ACTGCACTATAGCCTGGGCAACAGAGTGAGACTCTGTCTCGAAATAAAATTAAATTTAAAATTTAAAAAAGCTTCAAGGACAACCAGCAG ATGATGGCAGGACCAGGAAGGGTGCTTCAGGCAGCGGGAACTGAACCTGCACAGACTAGGGAAGCATGAACATCGGTACATCTGGGAGTG GCACAGCTTAGAGGGCCTGGAGCATGGAGTGTGAGGGGGAACTGGAGTGAGGGACAGGAGACCAGGCGACCATGGCCTTGCTGGGTAAGC GCTGTGACGTCCCCACCAACGGCTGCGGACCCGACCGCTGGAACTCCGCGTTCACCCGCAAAGACGAGATCATCACCAGCCTCGTGTCTG CCTTAGACTCCATGTGCTCAGCGCTGTCCAAACTGAACGCCGAGGTGGCCTGTGTCGCCGTGCACGATGAGAGCGCCTTTGTGGTGGGCA CAGAGAAGGGGAGAATGTTCCTGAATGCCCGGAAGGAGCTACAGTCAGACTTCCTCAGGTTCTGCCGAGGGCCCCCGTGGAAGGATCCGG AGGCAGAGCACCCCAAGAAGGTGCAGCGGGGCGAGGGTGGAGGCCGTAGCCTCCCTCGGTCCTCCCTGGAACATGGCTCAGATGTGTACC TTCTGCGGAAGATGGTAGAGGAGGTGTTTGATGTTCTTTATAGCGAGGCCCTGGGAAGGGCCAGTGTGGTGCCACTGCCCTATGAGAGGC TGCTCAGGGAGCCAGGGCTGCTGGCCGTGCAGGGGCTGCCCGAAGGCCTGGCCTTCCGAAGGCCAGCCGAGTATGACCCCAAGGCCCTCA TGGCCATCCTGGAACACAGCCACCGCATCCGCTTCAAGCTCAAGAGGCCACTTGAGGATGGCGGGCGGGACTCGAAGGCCCTGGTGGAGC TGAACGGTGTCTCCCTGATTCCCAAGGGGTCACGGGACTGTGGCCTGCATGGCCAGGCCCCCAAGGTGCCACCCCAGGACCTGCCCCCAA CCGCCACCTCCTCCTCCATGGCCAGCTTCCTGTACAGCACGGCGCTCCCCAACCACGCCATCCGAGAGCTCAAGCAGGAAGCACCTTCCT GCCCCCTTGCCCCCAGCGACCTGGGCCTGAGTCGGCCCATGCCAGAGCCCAAGGCCACCGGTGCCCAAGACTTCTCCGACTGTTGTGGAC AGAAGCCCACTGGGCCTGGTGGGCCTCTCATCCAGAACGTCCATGCCTCCAAGCGCATTCTCTTCTCCATCGTCCATGACAAGTCAGTGT ACCGCCGGAAGCACCAGGAGCTGCAAGCCATGCAGATGGAGCTGCAGAGCCCTGAGTACAAGCTGAGCAAGCTCCGCACCTCGACCATCA TGACCGACTACAACCCCAACTACTGCTTTGCTGGCAAGACCTCCTCCATCAGTGACCTGAAGGAGGTGCCGCGGAAAAACATCACCCTCA TTCGGGGTCTGGGCCATGGCGCCTTTGGGGAGGTGTATGAAGGCCAGGTGTCCGGAATGCCCAACGACCCAAGCCCCCTGCAAGTGGCTG TGAAGACGCTGCCTGAAGTGTGCTCTGAACAGGACGAACTGGATTTCCTCATGGAAGCCCTGATCATCAGCAAATTCAACCACCAGAACA TTGTTCGCTGCATTGGGGTGAGCCTGCAATCCCTGCCCCGGTTCATCCTGCTGGAGCTCATGGCGGGGGGAGACCTCAAGTCCTTCCTCC GAGAGACCCGCCCTCGCCCGAGCCAGCCCTCCTCCCTGGCCATGCTGGACCTTCTGCACGTGGCTCGGGACATTGCCTGTGGCTGTCAGT ATTTGGAGGAAAACCACTTCATCCACCGAGACATTGCTGCCAGAAACTGCCTCTTGACCTGTCCAGGCCCTGGAAGAGTGGCCAAGATTG GAGACTTCGGGATGGCCCGAGACATCTACAGGGCGAGCTACTATAGAAAGGGAGGCTGTGCCATGCTGCCAGTTAAGTGGATGCCCCCAG AGGCCTTCATGGAAGGAATATTCACTTCTAAAACAGACACATGGTCCTTTGGAGTGCTGCTATGGGAAATCTTTTCTCTTGGATATATGC CATACCCCAGCAAAAGCAACCAGGAAGTTCTGGAGTTTGTCACCAGTGGAGGCCGGATGGACCCACCCAAGAACTGCCCTGGGCCTGTAT ACCGGATAATGACTCAGTGCTGGCAACATCAGCCTGAAGACAGGCCCAACTTTGCCATCATTTTGGAGAGGATTGAATACTGCACCCAGG ACCCGGATGTAATCAACACCGCTTTGCCGATAGAATATGGTCCACTTGTGGAAGAGGAAGAGAAAGTGCCTGTGAGGCCCAAGGACCCTG AGGGGGTTCCTCCTCTCCTGGTCTCTCAACAGGCAAAACGGGAGGAGGAGCGCAGCCCAGCTGCCCCACCACCTCTGCCTACCACCTCCT CTGGCAAGGCTGCAAAGAAACCCACAGCTGCAGAGATCTCTGTTCGAGTCCCTAGAGGGCCGGCCGTGGAAGGGGGACACGTGAATATGG CATTCTCTCAGTCCAACCCTCCTTCGGAGTTGCACAAGGTCCACGGATCCAGAAACAAGCCCACCAGCTTGTGGAACCCAACGTACGGCT CCTGGTTTACAGAGAAACCCACCAAAAAGAATAATCCTATAGCAAAGAAGGAGCCACACGACAGGGGTAACCTGGGGCTGGAGGGAAGCT GTACTGTCCCACCTAACGTTGCAACTGGGAGACTTCCGGGGGCCTCACTGCTCCTAGAGCCCTCTTCGCTGACTGCCAATATGAAGGAGG TACCTCTGTTCAGGCTACGTCACTTCCCTTGTGGGAATGTCAATTACGGCTACCAGCAACAGGGCTTGCCCTTAGAAGCCGCTACTGCCC CTGGAGCTGGTCATTACGAGGATACCATTCTGAAAAGCAAGAATAGCATGAACCAGCCTGGGCCCTGAGCTCGGTCGCACACTCACTTCT CTTCCTTGGGATCCCTAAGACCGTGGAGGAGAGAGAGGCAATGGCTCCTTCACAAACCAGAGACCAAATGTCACGTTTTGTTTTGTGCCA ACCTATTTTGAAGTACCACCAAAAAAGCTGTATTTTGAAAATGCTTTAGAAAGGTTTTGAGCATGGGTTCATCCTATTCTTTCGAAAGAA GAAAATATCATAAAAATGAGTGATAAATACAAGGCCCAGATGTGGTTGCATAAGGTTTTTATGCATGTTTGTTGTATACTTCCTTATGCT TCTTTCAAATTGTGTGTGCTCTGCTTCAATGTAGTCAGAATTAGCTGCTTCTATGTTTCATAGTTGGGGTCATAGATGTTTCCTTGCCTT GTTGATGTGGACATGAGCCATTTGAGGGGAGAGGGAACGGAAATAAAGGAGTTATTTGTAATGACTAA >35245_35245_4_GTF2IRD1-ALK_GTF2IRD1_chr7_73935627_ENST00000476977_ALK_chr2_29446394_ENST00000389048_length(amino acids)=898AA_BP=335 MALLGKRCDVPTNGCGPDRWNSAFTRKDEIITSLVSALDSMCSALSKLNAEVACVAVHDESAFVVGTEKGRMFLNARKELQSDFLRFCRG PPWKDPEAEHPKKVQRGEGGGRSLPRSSLEHGSDVYLLRKMVEEVFDVLYSEALGRASVVPLPYERLLREPGLLAVQGLPEGLAFRRPAE YDPKALMAILEHSHRIRFKLKRPLEDGGRDSKALVELNGVSLIPKGSRDCGLHGQAPKVPPQDLPPTATSSSMASFLYSTALPNHAIREL KQEAPSCPLAPSDLGLSRPMPEPKATGAQDFSDCCGQKPTGPGGPLIQNVHASKRILFSIVHDKSVYRRKHQELQAMQMELQSPEYKLSK LRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIIS KFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGP GRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPK NCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPP PLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGN LGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQPGP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for GTF2IRD1-ALK |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for GTF2IRD1-ALK |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| Tgene | ALK | Q9UM73 | DB08865 | Crizotinib | Inhibitor | Small molecule | Approved |
| Tgene | ALK | Q9UM73 | DB09063 | Ceritinib | Antagonist | Small molecule | Approved |
| Tgene | ALK | Q9UM73 | DB11363 | Alectinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | ALK | Q9UM73 | DB12010 | Fostamatinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | ALK | Q9UM73 | DB12130 | Lorlatinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | ALK | Q9UM73 | DB12141 | Gilteritinib | Inhibitor | Small molecule | Approved|Investigational |
| Tgene | ALK | Q9UM73 | DB12267 | Brigatinib | Inhibitor | Small molecule | Approved|Investigational |
Top |
Related Diseases for GTF2IRD1-ALK |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | GTF2IRD1 | C0175702 | Williams Syndrome | 1 | CTD_human |
| Hgene | GTF2IRD1 | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Tgene | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human | |
| Tgene | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET | |
| Tgene | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human | |
| Tgene | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT | |
| Tgene | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human | |
| Tgene | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET | |
| Tgene | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human | |
| Tgene | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human | |
| Tgene | C0027627 | Neoplasm Metastasis | 2 | CTD_human | |
| Tgene | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET | |
| Tgene | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET | |
| Tgene | C0006118 | Brain Neoplasms | 1 | CGI;CTD_human | |
| Tgene | C0006142 | Malignant neoplasm of breast | 1 | CTD_human | |
| Tgene | C0007134 | Renal Cell Carcinoma | 1 | CTD_human | |
| Tgene | C0011570 | Mental Depression | 1 | PSYGENET | |
| Tgene | C0011581 | Depressive disorder | 1 | PSYGENET | |
| Tgene | C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human | |
| Tgene | C0036341 | Schizophrenia | 1 | PSYGENET | |
| Tgene | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human | |
| Tgene | C0085269 | Plasma Cell Granuloma, Pulmonary | 1 | CTD_human | |
| Tgene | C0153633 | Malignant neoplasm of brain | 1 | CGI;CTD_human | |
| Tgene | C0278601 | Inflammatory Breast Carcinoma | 1 | CTD_human | |
| Tgene | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human | |
| Tgene | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human | |
| Tgene | C0678222 | Breast Carcinoma | 1 | CTD_human | |
| Tgene | C0750974 | Brain Tumor, Primary | 1 | CTD_human | |
| Tgene | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human | |
| Tgene | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human | |
| Tgene | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human | |
| Tgene | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human | |
| Tgene | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human | |
| Tgene | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human | |
| Tgene | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human | |
| Tgene | C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 1 | ORPHANET | |
| Tgene | C1458155 | Mammary Neoplasms | 1 | CTD_human | |
| Tgene | C1527390 | Neoplasms, Intracranial | 1 | CTD_human | |
| Tgene | C2931189 | Neural crest tumor | 1 | ORPHANET | |
| Tgene | C3899155 | hereditary neuroblastoma | 1 | GENOMICS_ENGLAND | |
| Tgene | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
