
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:CDC42-LDLRAD2 (FusionGDB2 ID:HG998TG401944) |
Fusion Gene Summary for CDC42-LDLRAD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: CDC42-LDLRAD2 | Fusion gene ID: hg998tg401944 | Hgene | Tgene | Gene symbol | CDC42 | LDLRAD2 | Gene ID | 998 | 401944 |
| Gene name | cell division cycle 42 | low density lipoprotein receptor class A domain containing 2 | |
| Synonyms | CDC42Hs|G25K|TKS | - | |
| Cytomap | ('CDC42')('LDLRAD2') 1p36.12 | 1p36.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cell division control protein 42 homologG25K GTP-binding proteinGTP binding protein, 25kDadJ224A6.1.1 (cell division cycle 42 (GTP-binding protein, 25kD))dJ224A6.1.2 (cell division cycle 42 (GTP-binding protein, 25kD))growth-regulating proteinsmall | low-density lipoprotein receptor class A domain-containing protein 2low density lipoprotein receptor A domain containing 2 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | P60953 | Q5SZI1 | |
| Ensembl transtripts involved in fusion gene | ENST00000498236, ENST00000315554, ENST00000344548, ENST00000400259, ENST00000421089, | ||
| Fusion gene scores | * DoF score | 12 X 9 X 6=648 | 2 X 3 X 3=18 |
| # samples | 13 | 3 | |
| ** MAII score | log2(13/648*10)=-2.31748218985617 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: CDC42 [Title/Abstract] AND LDLRAD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | CDC42(22379235)-LDLRAD2(22138843), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CDC42-LDLRAD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42-LDLRAD2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42-LDLRAD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CDC42-LDLRAD2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CDC42 | GO:0030036 | actin cytoskeleton organization | 11035016 |
| Hgene | CDC42 | GO:0031274 | positive regulation of pseudopodium assembly | 11035016 |
| Hgene | CDC42 | GO:0051489 | regulation of filopodium assembly | 14978216 |
| Hgene | CDC42 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 11807099 |
 Fusion gene breakpoints across CDC42 (5'-gene) Fusion gene breakpoints across CDC42 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
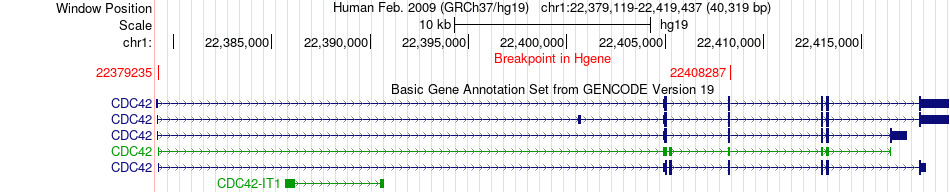 |
 Fusion gene breakpoints across LDLRAD2 (3'-gene) Fusion gene breakpoints across LDLRAD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
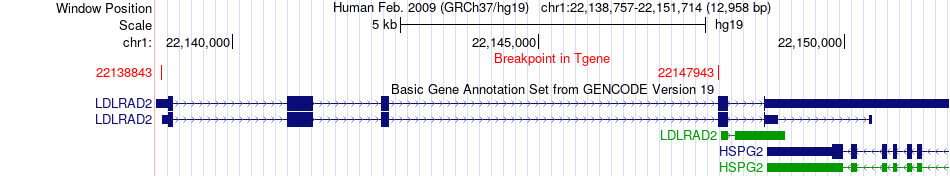 |
 Fusion gene information Fusion gene information * All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-1405-01A | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| ChimerDB4 | UCS | TCGA-N7-A59B | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| ChimerDB4 | UCS | TCGA-N8-A4PO | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
Top |
Fusion Gene ORF analysis for CDC42-LDLRAD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000498236 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 3UTR-3CDS | ENST00000498236 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 3UTR-5UTR | ENST00000498236 | ENST00000344642 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 3UTR-intron | ENST00000498236 | ENST00000484271 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 3UTR-intron | ENST00000498236 | ENST00000484271 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 3UTR-intron | ENST00000498236 | ENST00000543870 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5CDS-intron | ENST00000315554 | ENST00000484271 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 5CDS-intron | ENST00000344548 | ENST00000484271 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 5CDS-intron | ENST00000400259 | ENST00000484271 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 5CDS-intron | ENST00000421089 | ENST00000484271 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| 5UTR-5UTR | ENST00000315554 | ENST00000344642 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-5UTR | ENST00000344548 | ENST00000344642 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-5UTR | ENST00000400259 | ENST00000344642 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-5UTR | ENST00000421089 | ENST00000344642 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000315554 | ENST00000484271 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000315554 | ENST00000543870 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000344548 | ENST00000484271 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000344548 | ENST00000543870 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000400259 | ENST00000484271 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000400259 | ENST00000543870 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000421089 | ENST00000484271 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| 5UTR-intron | ENST00000421089 | ENST00000543870 | CDC42 | chr1 | 22379235 | + | LDLRAD2 | chr1 | 22138843 | + |
| In-frame | ENST00000315554 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000315554 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000344548 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000344548 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000400259 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000400259 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000421089 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
| In-frame | ENST00000421089 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000400259 | CDC42 | chr1 | 22408287 | + | ENST00000344642 | LDLRAD2 | chr1 | 22147943 | + | 3526 | 344 | 3131 | 2421 | 236 |
| ENST00000400259 | CDC42 | chr1 | 22408287 | + | ENST00000543870 | LDLRAD2 | chr1 | 22147943 | + | 792 | 344 | 660 | 160 | 166 |
| ENST00000344548 | CDC42 | chr1 | 22408287 | + | ENST00000344642 | LDLRAD2 | chr1 | 22147943 | + | 3611 | 429 | 3216 | 2506 | 236 |
| ENST00000344548 | CDC42 | chr1 | 22408287 | + | ENST00000543870 | LDLRAD2 | chr1 | 22147943 | + | 877 | 429 | 251 | 604 | 117 |
| ENST00000315554 | CDC42 | chr1 | 22408287 | + | ENST00000344642 | LDLRAD2 | chr1 | 22147943 | + | 3464 | 282 | 3069 | 2359 | 236 |
| ENST00000315554 | CDC42 | chr1 | 22408287 | + | ENST00000543870 | LDLRAD2 | chr1 | 22147943 | + | 730 | 282 | 104 | 457 | 117 |
| ENST00000421089 | CDC42 | chr1 | 22408287 | + | ENST00000344642 | LDLRAD2 | chr1 | 22147943 | + | 3618 | 436 | 3223 | 2513 | 236 |
| ENST00000421089 | CDC42 | chr1 | 22408287 | + | ENST00000543870 | LDLRAD2 | chr1 | 22147943 | + | 884 | 436 | 132 | 611 | 159 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000400259 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.5003174 | 0.4996826 |
| ENST00000400259 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.025317034 | 0.974683 |
| ENST00000344548 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.46547014 | 0.53452986 |
| ENST00000344548 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.013658943 | 0.98634106 |
| ENST00000315554 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.48021352 | 0.5197865 |
| ENST00000315554 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.015021463 | 0.9849785 |
| ENST00000421089 | ENST00000344642 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.49227756 | 0.5077225 |
| ENST00000421089 | ENST00000543870 | CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147943 | + | 0.02769202 | 0.97230804 |
Top |
Fusion Genomic Features for CDC42-LDLRAD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147942 | + | 0.000186616 | 0.9998134 |
| CDC42 | chr1 | 22408287 | + | LDLRAD2 | chr1 | 22147942 | + | 0.000186616 | 0.9998134 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
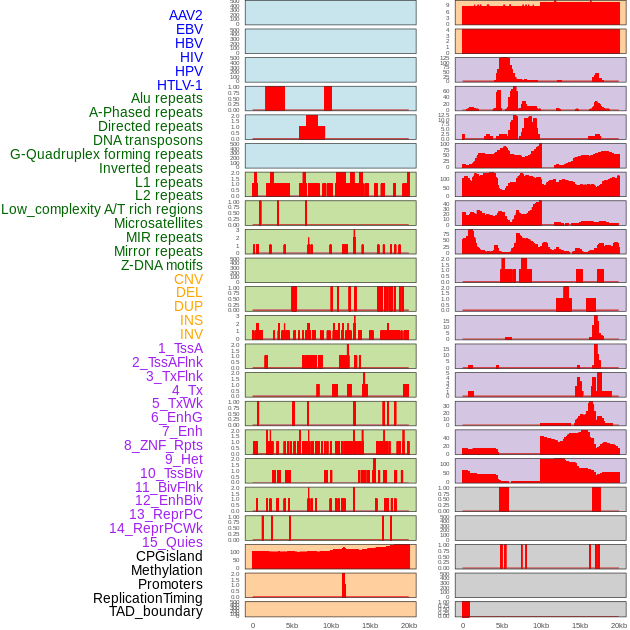 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
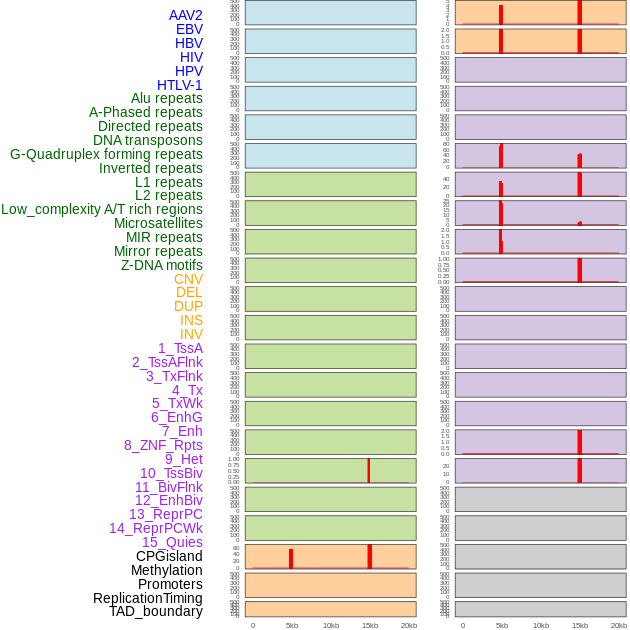 |
Top |
Fusion Protein Features for CDC42-LDLRAD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:22379235/chr1:22138843) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CDC42 | LDLRAD2 |
| FUNCTION: Plasma membrane-associated small GTPase which cycles between an active GTP-bound and an inactive GDP-bound state. In active state binds to a variety of effector proteins to regulate cellular responses. Involved in epithelial cell polarization processes. Regulates the bipolar attachment of spindle microtubules to kinetochores before chromosome congression in metaphase (PubMed:15642749). Regulates cell migration (PubMed:17038317). In neurons, plays a role in the extension and maintenance of the formation of filopodia, thin and actin-rich surface projections (PubMed:14978216). Required for DOCK10-mediated spine formation in Purkinje cells and hippocampal neurons. Facilitates filopodia formation upon DOCK11-activation (By similarity). Upon activation by CaMKII, modulates dendritic spine structural plasticity by relaying CaMKII transient activation to synapse-specific, long-term signaling (By similarity). Also plays a role in phagocytosis through organization of the F-actin cytoskeleton associated with forming phagocytic cups (PubMed:26465210). {ECO:0000250|UniProtKB:P60766, ECO:0000250|UniProtKB:Q8CFN2, ECO:0000269|PubMed:14978216, ECO:0000269|PubMed:15642749, ECO:0000269|PubMed:17038317, ECO:0000269|PubMed:26465210}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000315554 | + | 3 | 6 | 32_40 | 59 | 192.0 | Motif | Effector region |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000344548 | + | 4 | 7 | 32_40 | 59 | 192.0 | Motif | Effector region |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000400259 | + | 3 | 6 | 32_40 | 59 | 192.0 | Motif | Effector region |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000315554 | + | 3 | 6 | 10_17 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000315554 | + | 3 | 6 | 57_61 | 59 | 192.0 | Nucleotide binding | GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000344548 | + | 4 | 7 | 10_17 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000344548 | + | 4 | 7 | 57_61 | 59 | 192.0 | Nucleotide binding | GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000400259 | + | 3 | 6 | 10_17 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000400259 | + | 3 | 6 | 57_61 | 59 | 192.0 | Nucleotide binding | GTP |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000344642 | 2 | 5 | 251_272 | 214 | 273.0 | Topological domain | Cytoplasmic | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000543870 | 2 | 6 | 251_272 | 214 | 233.0 | Topological domain | Cytoplasmic | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000344642 | 2 | 5 | 234_250 | 214 | 273.0 | Transmembrane | Helical | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000543870 | 2 | 6 | 234_250 | 214 | 233.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000315554 | + | 3 | 6 | 115_118 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000344548 | + | 4 | 7 | 115_118 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Hgene | CDC42 | chr1:22408287 | chr1:22147943 | ENST00000400259 | + | 3 | 6 | 115_118 | 59 | 192.0 | Nucleotide binding | Note=GTP |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000344642 | 2 | 5 | 172_214 | 214 | 273.0 | Domain | LDL-receptor class A | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000543870 | 2 | 6 | 172_214 | 214 | 233.0 | Domain | LDL-receptor class A | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000344642 | 2 | 5 | 26_233 | 214 | 273.0 | Topological domain | Extracellular | |
| Tgene | LDLRAD2 | chr1:22408287 | chr1:22147943 | ENST00000543870 | 2 | 6 | 26_233 | 214 | 233.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for CDC42-LDLRAD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >14898_14898_1_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000315554_LDLRAD2_chr1_22147943_ENST00000344642_length(transcript)=3464nt_BP=282nt GGCAGCCGAGGAGACCCCGCGCAGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACA AAACTATTTCAGCAATGCAGACAATTAAGTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAA ACAAATTTCCATCGGAATATGTACCGACTGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTT TTGATACTGCAGGTCCCTCTCCGGTGCCCAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCG GGTCTGCAGGATCCCTCTGGATTGCAGCTGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCA CTGAGTGAAGCCCTCATCAAAGACTCAGGAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGG TCCCTTCTAGGACACAGAGGCCAGGCGTCCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCT GCAGATGGGGCAGGGTGGTCATATCCCCCTCCTCTCTCTCTCAGTCGTGAGTCCTGCCTTCCCCCACCTAAGGCACTAGCTCTTCCTGAG CACCAGCGGCATCCGTCCGTCCGTTGTCTGTTGGAGGAGTCCCTGGGCCTTCACTTCCAGATGGGTGGGGATGTGGCCTGGGGTGGGCGG GGCCCGGTGGGCGGGGCCCTATCAGTGCTGGGCTCTGCCTGTAGCAGCTCCTGGTAAGAGTTGGGGTGGGCCTTCCCTTACAGCCCCTGA GGGAGGGACCCCAGGCTGTGTGCTGGCAGGAGGGGGTGGGAATGCAGGCCAGGAGGACGTGGTAGGAGGGGGAGTCTGCCAGAGGAGGTG CCAGGGGCTGGGTGGAGGTGGGTGGGGGTGCTGAAAAACGAGCTGGTGGGGATGGGGACCGCCTGCCCAGGGGTGAGCTGCCTTTTGCTC CACAGCCGGCACTAAAGACAATTCCCAATCCTGAGTGGGTGGCAGAGACTCCTGCGATGCCCGTCTCAGGTAGCTGTGGGGCACCAGCCC ACAAGCCGAGGTTGGCTCTCCTAGGAGTGAGAACTGCCCAAGGGCTGCAGAAACAGGCCACCCAGCTCTATCTGGGGGCTCCATCGGTGG GTAGGGGGACAGTGGGGGCAGTTCTGGGCCCACCCAGCCACTGTTCCTGACCCCAAGTCCTGGTGACTTTCTGAGGTGCCCACTCCCATC CAACCTGCCTTGCTGGCCAGCCTTGTGGCTTTGCCCAGCTGTGTGTGTGAGGGTGGCATGCCCACCTCCAGTCCAGCCCAGGGCGGTAGC AGCAAAGCGTGGCATCGCCTCGGTTTCTTACAAAAATTCATAATAATATTAATAATAATATACTCGACATTGTCGGGCTGGGGCGTGGCC CGGGAGTCCGTGTGGGGCAGGCAGGTGCCTACGAGGGGCAGGGGCGTGTGTTGGCCCCGGCCTGGGCGCGGTGCTGCAGGTCCAGGGGCT GTGGGGGCGGGGCGCCGGGTCGGGCCGAGTGCAGCACCAGGTTCTTGACACAGCCTGTGATGCCTGAGGAGAATCTGCCCCCGGTCAGCG TGGCCACGTCAGGGGCTCCGCCTGCCGGGAGGTGAGAGGACAGGGCCTGTGGGCTCCAGCAGCCCAGGAGGCGAGGAAGGCTGGGCGAGC TCTAGCCCTAAGGGAGTGCCGTTCCTGCCCCTGCCCTGAGAAGGAGCCCCAGACTTACCGATGTAGACGCTGCCCTTGGCGTTGACTGCC ACGTTGGGACCTGGGGACCGGCCGCTGACCAGCTCCTCACCGTCGACTTGGATGGAACCTCTGCGGCCCTCCCTGCAGTGGAACTGGGTC AGGCCCCTTTCCACAAACTTCCTGGTCCTCCCCGGCCCCACGACAGAGTCCCCTCCCTCTGATATCGAGACTCCAGACTCAGAAGTCTGT CCCTGTTTCCCAAGCTCTTTCTTTCCCCCGCTGAACGAGAGATCGGGCCCCACAAACACAGCTTCCTCATTTCTGCACCCAGGTTTCCTC CCACTCCTGGGGCACTCGCCTGCCCCCAGCGGAGTTCAGTCCGAGATGGAAGCCCGAGCCCTGGCTGGTGGGTTCTCCCCTCCCCTGGCT TCAAGTTCTGTCTCCACAGAGCTCAATACCTGCCTCTCTGCCCATGGTAGGGGGCGTCCTGCCCCACTCCAGAACGCTGGGCCCCATCCC GAGTGCCCGGCAGGGTCCCTTACCGCAGTGCTGTCACCCGGTGCCACTCGCCGTCATTGATGGGGTCCTCAGAGACCAGGCGGGCCTCCC CACTACCCAGCTGGTACCTGCAGTCATCCAGGCCCAAGAAGTATGAGCTGGGGCAGGACCGGGGGGTGGGGTGCTGGGACCAGGGAAGGG AGAGGAAGGGCCAGGTGCCAGGACCTACCTGAAGACAAGGTGCCCGTCTTGAAGCCCGAGGCTGATGAAGTCCTTGCCTTGGCCGGCCTC TCCCACCTCCTGCCAGGGAAGCACAGGGTCTCTGGGGTCCCCAGCCTGGAGAGCAGAGGCTGCCGAGGCCAGGGGGCTCTGCTTTCCCCT CCCCCCACCACTCCGGCCACCAGGAAGCCAGCTTCCTGCCCCAGGAGCCCCAAGAGCCCAGCCGGATACCCACACTCACCACACCCTGCC AGAGCAGGAGGCCACTGGCTGTGCTGGTCCGAACCTCCAGCTCGATGGTCTCGGGCACCTCGGGCAGGCTGCGGAGGAAGAGCGGGTGAG GGGACAGAAGTCCCAGATTCCCATCCTCCCCATTAGGCCCATGGGCCCTTCCAATGCCAGTCTCACCTCCTGGAGAAGACATGGCCAGGG AAGGCGAGGAAGCCATCATCGTGGAAATAGGCTCCGTACTGCCCAGGGGCATCTGTGGGAGAGAGGAGGGTGGTGCCATACCTGCTGCAT CAGGCATCAAAATCCCCCGTCAGTTCCCCTGACCCCCACCTCCACGCCAACATGCAGGACTAGGGGGCTCCGAGCCTGCAGTCCCTGGGG ACCCACGGGGGCTGCCAACAGAATTCAGGGAGCCTATGACCTTGGATGGGAAAGCATTACACCTCAATTTCACTCACTTCTCACTGAAAT TGAGCAGTTCTTTCAATGAGATGTAGGCAAGAAACCAAAGTAGCAGTGGTAGGACCTGCAACTTTGTTACTAGTATTTGCTTATCACATG ACAGTAGCTGCAAATATTTCAAAATATCATTTACACATATTGCTGCTTTGAAATTGTGGTAGCTACTGAGATCACCAGATCTTGTTATTT GATGTGTTAATAAAGAAGCACATATATAGCAAGTCAGGAAACTT >14898_14898_1_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000315554_LDLRAD2_chr1_22147943_ENST00000344642_length(amino acids)=236AA_BP= MMPDAAGMAPPSSLPQMPLGSTEPISTMMASSPSLAMSSPGGETGIGRAHGPNGEDGNLGLLSPHPLFLRSLPEVPETIELEVRTSTASG LLLWQGVVSVGIRLGSWGSWGRKLASWWPEWWGEGKAEPPGLGSLCSPGWGPQRPCASLAGGGRGRPRQGLHQPRASRRAPCLQVGPGTW PFLSLPWSQHPTPRSCPSSYFLGLDDCRYQLGSGEARLVSEDPINDGEWHRVTALR -------------------------------------------------------------- >14898_14898_2_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000315554_LDLRAD2_chr1_22147943_ENST00000543870_length(transcript)=730nt_BP=282nt GGCAGCCGAGGAGACCCCGCGCAGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACA AAACTATTTCAGCAATGCAGACAATTAAGTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAA ACAAATTTCCATCGGAATATGTACCGACTGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTT TTGATACTGCAGGTCCCTCTCCGGTGCCCAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCG GGTCTGCAGGATCCCTCTGGATTGCAGCTGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCA CTGAGTGAAGCCCTCATCAAAGACTCAGGAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGG TCCCTTCTAGGACACAGAGGCCAGGCGTCCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCT GCAGATGGGGCAGGGTGGTCATATCCCCCTCCTCTCTCTCTCAGTCGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCAGCGGAGTT CAGTCCGAGA >14898_14898_2_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000315554_LDLRAD2_chr1_22147943_ENST00000543870_length(amino acids)=117AA_BP=58 MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGPSPVPSQTGSTDAHTSRSLTPSPALGSAGS LWIAAERSSPAGRDPTRQDAALEGSTE -------------------------------------------------------------- >14898_14898_3_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000344548_LDLRAD2_chr1_22147943_ENST00000344642_length(transcript)=3611nt_BP=429nt GTCAGGTGCGTGCCCCTGTCCGGCAGCCGAGGAGACCCCGCGCAGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGACGGAGTCTCACTGT GTTGCCCAGGCTGGAGTGCAGTGGCGCCATCTTGGCTCACTGCAGTGCGCCTCTGCCCCCCGAGTTCAAGCGATTCTCCTGCCTCAGGCT CCTGAGTAGCTGGGACTACAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGACAATTAAGTGTG TTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGTACCGACTGTTT TTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTGCCCAGCC AGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCAGCTGAGA GGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTCAGGAGGC CCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGCGTCCCAA CCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCCCCCTCCT CTCTCTCTCAGTCGTGAGTCCTGCCTTCCCCCACCTAAGGCACTAGCTCTTCCTGAGCACCAGCGGCATCCGTCCGTCCGTTGTCTGTTG GAGGAGTCCCTGGGCCTTCACTTCCAGATGGGTGGGGATGTGGCCTGGGGTGGGCGGGGCCCGGTGGGCGGGGCCCTATCAGTGCTGGGC TCTGCCTGTAGCAGCTCCTGGTAAGAGTTGGGGTGGGCCTTCCCTTACAGCCCCTGAGGGAGGGACCCCAGGCTGTGTGCTGGCAGGAGG GGGTGGGAATGCAGGCCAGGAGGACGTGGTAGGAGGGGGAGTCTGCCAGAGGAGGTGCCAGGGGCTGGGTGGAGGTGGGTGGGGGTGCTG AAAAACGAGCTGGTGGGGATGGGGACCGCCTGCCCAGGGGTGAGCTGCCTTTTGCTCCACAGCCGGCACTAAAGACAATTCCCAATCCTG AGTGGGTGGCAGAGACTCCTGCGATGCCCGTCTCAGGTAGCTGTGGGGCACCAGCCCACAAGCCGAGGTTGGCTCTCCTAGGAGTGAGAA CTGCCCAAGGGCTGCAGAAACAGGCCACCCAGCTCTATCTGGGGGCTCCATCGGTGGGTAGGGGGACAGTGGGGGCAGTTCTGGGCCCAC CCAGCCACTGTTCCTGACCCCAAGTCCTGGTGACTTTCTGAGGTGCCCACTCCCATCCAACCTGCCTTGCTGGCCAGCCTTGTGGCTTTG CCCAGCTGTGTGTGTGAGGGTGGCATGCCCACCTCCAGTCCAGCCCAGGGCGGTAGCAGCAAAGCGTGGCATCGCCTCGGTTTCTTACAA AAATTCATAATAATATTAATAATAATATACTCGACATTGTCGGGCTGGGGCGTGGCCCGGGAGTCCGTGTGGGGCAGGCAGGTGCCTACG AGGGGCAGGGGCGTGTGTTGGCCCCGGCCTGGGCGCGGTGCTGCAGGTCCAGGGGCTGTGGGGGCGGGGCGCCGGGTCGGGCCGAGTGCA GCACCAGGTTCTTGACACAGCCTGTGATGCCTGAGGAGAATCTGCCCCCGGTCAGCGTGGCCACGTCAGGGGCTCCGCCTGCCGGGAGGT GAGAGGACAGGGCCTGTGGGCTCCAGCAGCCCAGGAGGCGAGGAAGGCTGGGCGAGCTCTAGCCCTAAGGGAGTGCCGTTCCTGCCCCTG CCCTGAGAAGGAGCCCCAGACTTACCGATGTAGACGCTGCCCTTGGCGTTGACTGCCACGTTGGGACCTGGGGACCGGCCGCTGACCAGC TCCTCACCGTCGACTTGGATGGAACCTCTGCGGCCCTCCCTGCAGTGGAACTGGGTCAGGCCCCTTTCCACAAACTTCCTGGTCCTCCCC GGCCCCACGACAGAGTCCCCTCCCTCTGATATCGAGACTCCAGACTCAGAAGTCTGTCCCTGTTTCCCAAGCTCTTTCTTTCCCCCGCTG AACGAGAGATCGGGCCCCACAAACACAGCTTCCTCATTTCTGCACCCAGGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCAGCGGA GTTCAGTCCGAGATGGAAGCCCGAGCCCTGGCTGGTGGGTTCTCCCCTCCCCTGGCTTCAAGTTCTGTCTCCACAGAGCTCAATACCTGC CTCTCTGCCCATGGTAGGGGGCGTCCTGCCCCACTCCAGAACGCTGGGCCCCATCCCGAGTGCCCGGCAGGGTCCCTTACCGCAGTGCTG TCACCCGGTGCCACTCGCCGTCATTGATGGGGTCCTCAGAGACCAGGCGGGCCTCCCCACTACCCAGCTGGTACCTGCAGTCATCCAGGC CCAAGAAGTATGAGCTGGGGCAGGACCGGGGGGTGGGGTGCTGGGACCAGGGAAGGGAGAGGAAGGGCCAGGTGCCAGGACCTACCTGAA GACAAGGTGCCCGTCTTGAAGCCCGAGGCTGATGAAGTCCTTGCCTTGGCCGGCCTCTCCCACCTCCTGCCAGGGAAGCACAGGGTCTCT GGGGTCCCCAGCCTGGAGAGCAGAGGCTGCCGAGGCCAGGGGGCTCTGCTTTCCCCTCCCCCCACCACTCCGGCCACCAGGAAGCCAGCT TCCTGCCCCAGGAGCCCCAAGAGCCCAGCCGGATACCCACACTCACCACACCCTGCCAGAGCAGGAGGCCACTGGCTGTGCTGGTCCGAA CCTCCAGCTCGATGGTCTCGGGCACCTCGGGCAGGCTGCGGAGGAAGAGCGGGTGAGGGGACAGAAGTCCCAGATTCCCATCCTCCCCAT TAGGCCCATGGGCCCTTCCAATGCCAGTCTCACCTCCTGGAGAAGACATGGCCAGGGAAGGCGAGGAAGCCATCATCGTGGAAATAGGCT CCGTACTGCCCAGGGGCATCTGTGGGAGAGAGGAGGGTGGTGCCATACCTGCTGCATCAGGCATCAAAATCCCCCGTCAGTTCCCCTGAC CCCCACCTCCACGCCAACATGCAGGACTAGGGGGCTCCGAGCCTGCAGTCCCTGGGGACCCACGGGGGCTGCCAACAGAATTCAGGGAGC CTATGACCTTGGATGGGAAAGCATTACACCTCAATTTCACTCACTTCTCACTGAAATTGAGCAGTTCTTTCAATGAGATGTAGGCAAGAA ACCAAAGTAGCAGTGGTAGGACCTGCAACTTTGTTACTAGTATTTGCTTATCACATGACAGTAGCTGCAAATATTTCAAAATATCATTTA CACATATTGCTGCTTTGAAATTGTGGTAGCTACTGAGATCACCAGATCTTGTTATTTGATGTGTTAATAAAGAAGCACATATATAGCAAG TCAGGAAACTT >14898_14898_3_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000344548_LDLRAD2_chr1_22147943_ENST00000344642_length(amino acids)=236AA_BP= MMPDAAGMAPPSSLPQMPLGSTEPISTMMASSPSLAMSSPGGETGIGRAHGPNGEDGNLGLLSPHPLFLRSLPEVPETIELEVRTSTASG LLLWQGVVSVGIRLGSWGSWGRKLASWWPEWWGEGKAEPPGLGSLCSPGWGPQRPCASLAGGGRGRPRQGLHQPRASRRAPCLQVGPGTW PFLSLPWSQHPTPRSCPSSYFLGLDDCRYQLGSGEARLVSEDPINDGEWHRVTALR -------------------------------------------------------------- >14898_14898_4_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000344548_LDLRAD2_chr1_22147943_ENST00000543870_length(transcript)=877nt_BP=429nt GTCAGGTGCGTGCCCCTGTCCGGCAGCCGAGGAGACCCCGCGCAGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGACGGAGTCTCACTGT GTTGCCCAGGCTGGAGTGCAGTGGCGCCATCTTGGCTCACTGCAGTGCGCCTCTGCCCCCCGAGTTCAAGCGATTCTCCTGCCTCAGGCT CCTGAGTAGCTGGGACTACAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGACAATTAAGTGTG TTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGTACCGACTGTTT TTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTGCCCAGCC AGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCAGCTGAGA GGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTCAGGAGGC CCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGCGTCCCAA CCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCCCCCTCCT CTCTCTCTCAGTCGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCAGCGGAGTTCAGTCCGAGA >14898_14898_4_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000344548_LDLRAD2_chr1_22147943_ENST00000543870_length(amino acids)=117AA_BP=58 MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGPSPVPSQTGSTDAHTSRSLTPSPALGSAGS LWIAAERSSPAGRDPTRQDAALEGSTE -------------------------------------------------------------- >14898_14898_5_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000400259_LDLRAD2_chr1_22147943_ENST00000344642_length(transcript)=3526nt_BP=344nt ACTTCCGCGGGCACCCAACTGTGCGTCTCCTGCGCGCTGACGTCAGGTGCGTGCCCCTGTCCGGCAGCCGAGGAGACCCCGCGCAGTGCT GCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGACAATTAA GTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGTACCGAC TGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTGCC CAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCAGC TGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTCAG GAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGCGT CCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCCCC CTCCTCTCTCTCTCAGTCGTGAGTCCTGCCTTCCCCCACCTAAGGCACTAGCTCTTCCTGAGCACCAGCGGCATCCGTCCGTCCGTTGTC TGTTGGAGGAGTCCCTGGGCCTTCACTTCCAGATGGGTGGGGATGTGGCCTGGGGTGGGCGGGGCCCGGTGGGCGGGGCCCTATCAGTGC TGGGCTCTGCCTGTAGCAGCTCCTGGTAAGAGTTGGGGTGGGCCTTCCCTTACAGCCCCTGAGGGAGGGACCCCAGGCTGTGTGCTGGCA GGAGGGGGTGGGAATGCAGGCCAGGAGGACGTGGTAGGAGGGGGAGTCTGCCAGAGGAGGTGCCAGGGGCTGGGTGGAGGTGGGTGGGGG TGCTGAAAAACGAGCTGGTGGGGATGGGGACCGCCTGCCCAGGGGTGAGCTGCCTTTTGCTCCACAGCCGGCACTAAAGACAATTCCCAA TCCTGAGTGGGTGGCAGAGACTCCTGCGATGCCCGTCTCAGGTAGCTGTGGGGCACCAGCCCACAAGCCGAGGTTGGCTCTCCTAGGAGT GAGAACTGCCCAAGGGCTGCAGAAACAGGCCACCCAGCTCTATCTGGGGGCTCCATCGGTGGGTAGGGGGACAGTGGGGGCAGTTCTGGG CCCACCCAGCCACTGTTCCTGACCCCAAGTCCTGGTGACTTTCTGAGGTGCCCACTCCCATCCAACCTGCCTTGCTGGCCAGCCTTGTGG CTTTGCCCAGCTGTGTGTGTGAGGGTGGCATGCCCACCTCCAGTCCAGCCCAGGGCGGTAGCAGCAAAGCGTGGCATCGCCTCGGTTTCT TACAAAAATTCATAATAATATTAATAATAATATACTCGACATTGTCGGGCTGGGGCGTGGCCCGGGAGTCCGTGTGGGGCAGGCAGGTGC CTACGAGGGGCAGGGGCGTGTGTTGGCCCCGGCCTGGGCGCGGTGCTGCAGGTCCAGGGGCTGTGGGGGCGGGGCGCCGGGTCGGGCCGA GTGCAGCACCAGGTTCTTGACACAGCCTGTGATGCCTGAGGAGAATCTGCCCCCGGTCAGCGTGGCCACGTCAGGGGCTCCGCCTGCCGG GAGGTGAGAGGACAGGGCCTGTGGGCTCCAGCAGCCCAGGAGGCGAGGAAGGCTGGGCGAGCTCTAGCCCTAAGGGAGTGCCGTTCCTGC CCCTGCCCTGAGAAGGAGCCCCAGACTTACCGATGTAGACGCTGCCCTTGGCGTTGACTGCCACGTTGGGACCTGGGGACCGGCCGCTGA CCAGCTCCTCACCGTCGACTTGGATGGAACCTCTGCGGCCCTCCCTGCAGTGGAACTGGGTCAGGCCCCTTTCCACAAACTTCCTGGTCC TCCCCGGCCCCACGACAGAGTCCCCTCCCTCTGATATCGAGACTCCAGACTCAGAAGTCTGTCCCTGTTTCCCAAGCTCTTTCTTTCCCC CGCTGAACGAGAGATCGGGCCCCACAAACACAGCTTCCTCATTTCTGCACCCAGGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCA GCGGAGTTCAGTCCGAGATGGAAGCCCGAGCCCTGGCTGGTGGGTTCTCCCCTCCCCTGGCTTCAAGTTCTGTCTCCACAGAGCTCAATA CCTGCCTCTCTGCCCATGGTAGGGGGCGTCCTGCCCCACTCCAGAACGCTGGGCCCCATCCCGAGTGCCCGGCAGGGTCCCTTACCGCAG TGCTGTCACCCGGTGCCACTCGCCGTCATTGATGGGGTCCTCAGAGACCAGGCGGGCCTCCCCACTACCCAGCTGGTACCTGCAGTCATC CAGGCCCAAGAAGTATGAGCTGGGGCAGGACCGGGGGGTGGGGTGCTGGGACCAGGGAAGGGAGAGGAAGGGCCAGGTGCCAGGACCTAC CTGAAGACAAGGTGCCCGTCTTGAAGCCCGAGGCTGATGAAGTCCTTGCCTTGGCCGGCCTCTCCCACCTCCTGCCAGGGAAGCACAGGG TCTCTGGGGTCCCCAGCCTGGAGAGCAGAGGCTGCCGAGGCCAGGGGGCTCTGCTTTCCCCTCCCCCCACCACTCCGGCCACCAGGAAGC CAGCTTCCTGCCCCAGGAGCCCCAAGAGCCCAGCCGGATACCCACACTCACCACACCCTGCCAGAGCAGGAGGCCACTGGCTGTGCTGGT CCGAACCTCCAGCTCGATGGTCTCGGGCACCTCGGGCAGGCTGCGGAGGAAGAGCGGGTGAGGGGACAGAAGTCCCAGATTCCCATCCTC CCCATTAGGCCCATGGGCCCTTCCAATGCCAGTCTCACCTCCTGGAGAAGACATGGCCAGGGAAGGCGAGGAAGCCATCATCGTGGAAAT AGGCTCCGTACTGCCCAGGGGCATCTGTGGGAGAGAGGAGGGTGGTGCCATACCTGCTGCATCAGGCATCAAAATCCCCCGTCAGTTCCC CTGACCCCCACCTCCACGCCAACATGCAGGACTAGGGGGCTCCGAGCCTGCAGTCCCTGGGGACCCACGGGGGCTGCCAACAGAATTCAG GGAGCCTATGACCTTGGATGGGAAAGCATTACACCTCAATTTCACTCACTTCTCACTGAAATTGAGCAGTTCTTTCAATGAGATGTAGGC AAGAAACCAAAGTAGCAGTGGTAGGACCTGCAACTTTGTTACTAGTATTTGCTTATCACATGACAGTAGCTGCAAATATTTCAAAATATC ATTTACACATATTGCTGCTTTGAAATTGTGGTAGCTACTGAGATCACCAGATCTTGTTATTTGATGTGTTAATAAAGAAGCACATATATA GCAAGTCAGGAAACTT >14898_14898_5_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000400259_LDLRAD2_chr1_22147943_ENST00000344642_length(amino acids)=236AA_BP= MMPDAAGMAPPSSLPQMPLGSTEPISTMMASSPSLAMSSPGGETGIGRAHGPNGEDGNLGLLSPHPLFLRSLPEVPETIELEVRTSTASG LLLWQGVVSVGIRLGSWGSWGRKLASWWPEWWGEGKAEPPGLGSLCSPGWGPQRPCASLAGGGRGRPRQGLHQPRASRRAPCLQVGPGTW PFLSLPWSQHPTPRSCPSSYFLGLDDCRYQLGSGEARLVSEDPINDGEWHRVTALR -------------------------------------------------------------- >14898_14898_6_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000400259_LDLRAD2_chr1_22147943_ENST00000543870_length(transcript)=792nt_BP=344nt ACTTCCGCGGGCACCCAACTGTGCGTCTCCTGCGCGCTGACGTCAGGTGCGTGCCCCTGTCCGGCAGCCGAGGAGACCCCGCGCAGTGCT GCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGACAATTAA GTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGTACCGAC TGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTGCC CAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCAGC TGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTCAG GAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGCGT CCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCCCC CTCCTCTCTCTCTCAGTCGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCAGCGGAGTTCAGTCCGAGA >14898_14898_6_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000400259_LDLRAD2_chr1_22147943_ENST00000543870_length(amino acids)=166AA_BP=1 MPVAPRLWGWDAWPLCPRRDPPVVFVLIFLNKRCYPRQGPPESLMRASLSGAFQSCVLSRGVPACWGTPLSCNPEGSCRPESWGGSQGSG GMGICTSCLAGHRRGTCSIKKSKSIWFSTNHNCDCIVVKNSRYIFRWKFVCCVGYQETCFTNSTIAHNNTLNCLHC -------------------------------------------------------------- >14898_14898_7_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000421089_LDLRAD2_chr1_22147943_ENST00000344642_length(transcript)=3618nt_BP=436nt AGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGAC AATTAAGTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGT ACCGACTATCTTGACTTCTCATGGGTAAATTATATACACTTTAAACAGCTGAAAAATCAGTGGAAAGTCAGAAGGGGTGACACAGGGTTT GCAAGAAGTGCTGGGAGGCAAAACTCCAGTAGACAAGATTCTAACGAGTGGTGGTCTCAATTTGGTGAAGTATGCCCTACATCTTGGAAT GAGGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTG CCCAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCA GCTGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTC AGGAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGC GTCCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCC CCCTCCTCTCTCTCTCAGTCGTGAGTCCTGCCTTCCCCCACCTAAGGCACTAGCTCTTCCTGAGCACCAGCGGCATCCGTCCGTCCGTTG TCTGTTGGAGGAGTCCCTGGGCCTTCACTTCCAGATGGGTGGGGATGTGGCCTGGGGTGGGCGGGGCCCGGTGGGCGGGGCCCTATCAGT GCTGGGCTCTGCCTGTAGCAGCTCCTGGTAAGAGTTGGGGTGGGCCTTCCCTTACAGCCCCTGAGGGAGGGACCCCAGGCTGTGTGCTGG CAGGAGGGGGTGGGAATGCAGGCCAGGAGGACGTGGTAGGAGGGGGAGTCTGCCAGAGGAGGTGCCAGGGGCTGGGTGGAGGTGGGTGGG GGTGCTGAAAAACGAGCTGGTGGGGATGGGGACCGCCTGCCCAGGGGTGAGCTGCCTTTTGCTCCACAGCCGGCACTAAAGACAATTCCC AATCCTGAGTGGGTGGCAGAGACTCCTGCGATGCCCGTCTCAGGTAGCTGTGGGGCACCAGCCCACAAGCCGAGGTTGGCTCTCCTAGGA GTGAGAACTGCCCAAGGGCTGCAGAAACAGGCCACCCAGCTCTATCTGGGGGCTCCATCGGTGGGTAGGGGGACAGTGGGGGCAGTTCTG GGCCCACCCAGCCACTGTTCCTGACCCCAAGTCCTGGTGACTTTCTGAGGTGCCCACTCCCATCCAACCTGCCTTGCTGGCCAGCCTTGT GGCTTTGCCCAGCTGTGTGTGTGAGGGTGGCATGCCCACCTCCAGTCCAGCCCAGGGCGGTAGCAGCAAAGCGTGGCATCGCCTCGGTTT CTTACAAAAATTCATAATAATATTAATAATAATATACTCGACATTGTCGGGCTGGGGCGTGGCCCGGGAGTCCGTGTGGGGCAGGCAGGT GCCTACGAGGGGCAGGGGCGTGTGTTGGCCCCGGCCTGGGCGCGGTGCTGCAGGTCCAGGGGCTGTGGGGGCGGGGCGCCGGGTCGGGCC GAGTGCAGCACCAGGTTCTTGACACAGCCTGTGATGCCTGAGGAGAATCTGCCCCCGGTCAGCGTGGCCACGTCAGGGGCTCCGCCTGCC GGGAGGTGAGAGGACAGGGCCTGTGGGCTCCAGCAGCCCAGGAGGCGAGGAAGGCTGGGCGAGCTCTAGCCCTAAGGGAGTGCCGTTCCT GCCCCTGCCCTGAGAAGGAGCCCCAGACTTACCGATGTAGACGCTGCCCTTGGCGTTGACTGCCACGTTGGGACCTGGGGACCGGCCGCT GACCAGCTCCTCACCGTCGACTTGGATGGAACCTCTGCGGCCCTCCCTGCAGTGGAACTGGGTCAGGCCCCTTTCCACAAACTTCCTGGT CCTCCCCGGCCCCACGACAGAGTCCCCTCCCTCTGATATCGAGACTCCAGACTCAGAAGTCTGTCCCTGTTTCCCAAGCTCTTTCTTTCC CCCGCTGAACGAGAGATCGGGCCCCACAAACACAGCTTCCTCATTTCTGCACCCAGGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCC CAGCGGAGTTCAGTCCGAGATGGAAGCCCGAGCCCTGGCTGGTGGGTTCTCCCCTCCCCTGGCTTCAAGTTCTGTCTCCACAGAGCTCAA TACCTGCCTCTCTGCCCATGGTAGGGGGCGTCCTGCCCCACTCCAGAACGCTGGGCCCCATCCCGAGTGCCCGGCAGGGTCCCTTACCGC AGTGCTGTCACCCGGTGCCACTCGCCGTCATTGATGGGGTCCTCAGAGACCAGGCGGGCCTCCCCACTACCCAGCTGGTACCTGCAGTCA TCCAGGCCCAAGAAGTATGAGCTGGGGCAGGACCGGGGGGTGGGGTGCTGGGACCAGGGAAGGGAGAGGAAGGGCCAGGTGCCAGGACCT ACCTGAAGACAAGGTGCCCGTCTTGAAGCCCGAGGCTGATGAAGTCCTTGCCTTGGCCGGCCTCTCCCACCTCCTGCCAGGGAAGCACAG GGTCTCTGGGGTCCCCAGCCTGGAGAGCAGAGGCTGCCGAGGCCAGGGGGCTCTGCTTTCCCCTCCCCCCACCACTCCGGCCACCAGGAA GCCAGCTTCCTGCCCCAGGAGCCCCAAGAGCCCAGCCGGATACCCACACTCACCACACCCTGCCAGAGCAGGAGGCCACTGGCTGTGCTG GTCCGAACCTCCAGCTCGATGGTCTCGGGCACCTCGGGCAGGCTGCGGAGGAAGAGCGGGTGAGGGGACAGAAGTCCCAGATTCCCATCC TCCCCATTAGGCCCATGGGCCCTTCCAATGCCAGTCTCACCTCCTGGAGAAGACATGGCCAGGGAAGGCGAGGAAGCCATCATCGTGGAA ATAGGCTCCGTACTGCCCAGGGGCATCTGTGGGAGAGAGGAGGGTGGTGCCATACCTGCTGCATCAGGCATCAAAATCCCCCGTCAGTTC CCCTGACCCCCACCTCCACGCCAACATGCAGGACTAGGGGGCTCCGAGCCTGCAGTCCCTGGGGACCCACGGGGGCTGCCAACAGAATTC AGGGAGCCTATGACCTTGGATGGGAAAGCATTACACCTCAATTTCACTCACTTCTCACTGAAATTGAGCAGTTCTTTCAATGAGATGTAG GCAAGAAACCAAAGTAGCAGTGGTAGGACCTGCAACTTTGTTACTAGTATTTGCTTATCACATGACAGTAGCTGCAAATATTTCAAAATA TCATTTACACATATTGCTGCTTTGAAATTGTGGTAGCTACTGAGATCACCAGATCTTGTTATTTGATGTGTTAATAAAGAAGCACATATA TAGCAAGTCAGGAAACTT >14898_14898_7_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000421089_LDLRAD2_chr1_22147943_ENST00000344642_length(amino acids)=236AA_BP= MMPDAAGMAPPSSLPQMPLGSTEPISTMMASSPSLAMSSPGGETGIGRAHGPNGEDGNLGLLSPHPLFLRSLPEVPETIELEVRTSTASG LLLWQGVVSVGIRLGSWGSWGRKLASWWPEWWGEGKAEPPGLGSLCSPGWGPQRPCASLAGGGRGRPRQGLHQPRASRRAPCLQVGPGTW PFLSLPWSQHPTPRSCPSSYFLGLDDCRYQLGSGEARLVSEDPINDGEWHRVTALR -------------------------------------------------------------- >14898_14898_8_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000421089_LDLRAD2_chr1_22147943_ENST00000543870_length(transcript)=884nt_BP=436nt AGTGCTGCCAACGCCCCGGTGGAGAAGCTGAGGTCATCATCAGATTTGAAATATTTAAAGTGGATACAAAACTATTTCAGCAATGCAGAC AATTAAGTGTGTTGTTGTGGGCGATGGTGCTGTTGGTAAAACATGTCTCCTGATATCCTACACAACAAACAAATTTCCATCGGAATATGT ACCGACTATCTTGACTTCTCATGGGTAAATTATATACACTTTAAACAGCTGAAAAATCAGTGGAAAGTCAGAAGGGGTGACACAGGGTTT GCAAGAAGTGCTGGGAGGCAAAACTCCAGTAGACAAGATTCTAACGAGTGGTGGTCTCAATTTGGTGAAGTATGCCCTACATCTTGGAAT GAGGTTTTTGACAACTATGCAGTCACAGTTATGATTGGTGGAGAACCATATACTCTTGGACTTTTTGATACTGCAGGTCCCTCTCCGGTG CCCAGCCAGACAGGAAGTACAGATGCCCATACCTCCAGATCCCTGACTCCCTCCCCAGCTCTCGGGTCTGCAGGATCCCTCTGGATTGCA GCTGAGAGGAGTTCCCCAGCAGGCAGGGACCCCACGAGACAAGACGCAGCTTTGGAAGGCTCCACTGAGTGAAGCCCTCATCAAAGACTC AGGAGGCCCCTGGCGGGGATAGCACCGTTTATTAAGAAAAATCAAGACAAAGACCACAGGAGGGTCCCTTCTAGGACACAGAGGCCAGGC GTCCCAACCCCACAGTCTGGGGGCCACTGGCAGGATGGCACTTGAGCTGGATCTTCAGGCTCCTGCAGATGGGGCAGGGTGGTCATATCC CCCTCCTCTCTCTCTCAGTCGTTTCCTCCCACTCCTGGGGCACTCGCCTGCCCCCAGCGGAGTTCAGTCCGAGA >14898_14898_8_CDC42-LDLRAD2_CDC42_chr1_22408287_ENST00000421089_LDLRAD2_chr1_22147943_ENST00000543870_length(amino acids)=159AA_BP=100 MSPDILHNKQISIGICTDYLDFSWVNYIHFKQLKNQWKVRRGDTGFARSAGRQNSSRQDSNEWWSQFGEVCPTSWNEVFDNYAVTVMIGG EPYTLGLFDTAGPSPVPSQTGSTDAHTSRSLTPSPALGSAGSLWIAAERSSPAGRDPTRQDAALEGSTE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for CDC42-LDLRAD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for CDC42-LDLRAD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for CDC42-LDLRAD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CDC42 | C0036341 | Schizophrenia | 4 | PSYGENET |
| Hgene | CDC42 | C4225222 | TAKENOUCHI-KOSAKI SYNDROME | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | CDC42 | C0001624 | Adrenal Gland Neoplasms | 1 | CTD_human |
| Hgene | CDC42 | C0005818 | Blood Platelet Disorders | 1 | GENOMICS_ENGLAND |
| Hgene | CDC42 | C0007097 | Carcinoma | 1 | CTD_human |
| Hgene | CDC42 | C0019693 | HIV Infections | 1 | CTD_human |
| Hgene | CDC42 | C0024667 | Animal Mammary Neoplasms | 1 | CTD_human |
| Hgene | CDC42 | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| Hgene | CDC42 | C0027540 | Necrosis | 1 | CTD_human |
| Hgene | CDC42 | C0205696 | Anaplastic carcinoma | 1 | CTD_human |
| Hgene | CDC42 | C0205697 | Carcinoma, Spindle-Cell | 1 | CTD_human |
| Hgene | CDC42 | C0205698 | Undifferentiated carcinoma | 1 | CTD_human |
| Hgene | CDC42 | C0205699 | Carcinomatosis | 1 | CTD_human |
| Hgene | CDC42 | C0750887 | Adrenal Cancer | 1 | CTD_human |
| Hgene | CDC42 | C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human |
| Hgene | CDC42 | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |
| Hgene | CDC42 | C4505456 | HIV Coinfection | 1 | CTD_human |
