| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CUL2-PARD3 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CUL2-PARD3 | FusionPDB ID: 20659 | FusionGDB2.0 ID: 20659 | Hgene | Tgene | Gene symbol | CUL2 | PARD3 | Gene ID | 8453 | 56288 |
| Gene name | cullin 2 | par-3 family cell polarity regulator | |
| Synonyms | - | ASIP|Baz|PAR3|PAR3alpha|PARD-3|PARD3A|PPP1R118|SE2-5L16|SE2-5LT1|SE2-5T2 | |
| Cytomap | 10p11.21 | 10p11.22-p11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cullin-2CUL-2testis secretory sperm-binding protein Li 238E | partitioning defective 3 homologCTCL tumor antigen se2-5PAR3-alphaatypical PKC isotype-specific interacting proteinbazookapar-3 family cell polarity regulator alphapar-3 partitioning defective 3 homologprotein phosphatase 1, regulatory subunit 118 | |
| Modification date | 20200327 | 20200327 | |
| UniProtAcc | Q13617 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000374742, ENST00000374746, ENST00000374748, ENST00000374749, ENST00000374751, ENST00000537177, ENST00000602371, ENST00000478044, | ENST00000340077, ENST00000374768, ENST00000374773, ENST00000374776, ENST00000466092, ENST00000544292, ENST00000346874, ENST00000350537, ENST00000374788, ENST00000374789, ENST00000374790, ENST00000374794, ENST00000545260, ENST00000545693, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 7 X 7 X 5=245 | 20 X 21 X 8=3360 |
| # samples | 8 | 24 | |
| ** MAII score | log2(8/245*10)=-1.61470984411521 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(24/3360*10)=-3.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CUL2 [Title/Abstract] AND PARD3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CUL2(35333494)-PARD3(34806087), # samples:2 PARD3(35103803)-CUL2(35360267), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CUL2-PARD3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CUL2-PARD3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PARD3-CUL2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PARD3-CUL2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CUL2-PARD3 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. CUL2-PARD3 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CUL2-PARD3 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. CUL2-PARD3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. PARD3-CUL2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. PARD3-CUL2 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. PARD3-CUL2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. PARD3-CUL2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across CUL2 (5'-gene) Fusion gene breakpoints across CUL2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
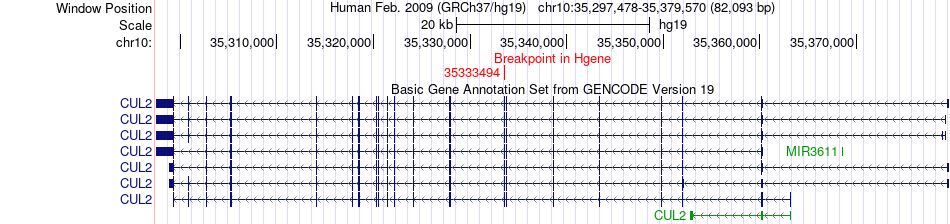 |
 Fusion gene breakpoints across PARD3 (3'-gene) Fusion gene breakpoints across PARD3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
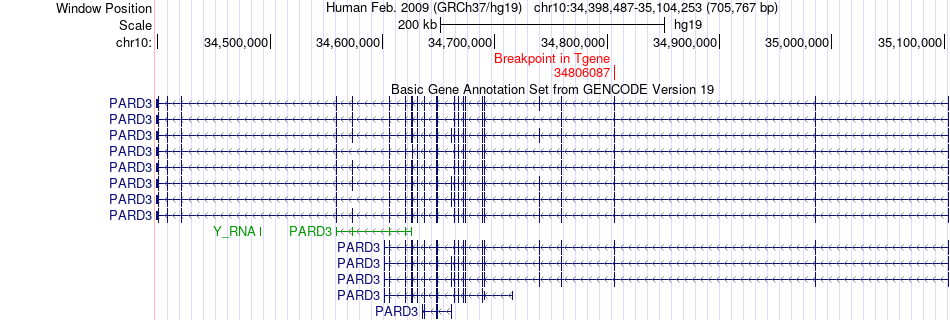 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-6642-01A | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000545260 | PARD3 | chr10 | 34806087 | - | 5924 | 736 | 22 | 4314 | 1430 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000545693 | PARD3 | chr10 | 34806087 | - | 6146 | 736 | 22 | 4536 | 1504 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000350537 | PARD3 | chr10 | 34806087 | - | 6055 | 736 | 22 | 4446 | 1474 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000346874 | PARD3 | chr10 | 34806087 | - | 6082 | 736 | 22 | 4473 | 1483 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000374789 | PARD3 | chr10 | 34806087 | - | 6193 | 736 | 22 | 4584 | 1520 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000374790 | PARD3 | chr10 | 34806087 | - | 6013 | 736 | 22 | 4404 | 1460 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000374794 | PARD3 | chr10 | 34806087 | - | 5857 | 736 | 22 | 4248 | 1408 |
| ENST00000374746 | CUL2 | chr10 | 35333494 | - | ENST00000374788 | PARD3 | chr10 | 34806087 | - | 6184 | 736 | 22 | 4575 | 1517 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000545260 | PARD3 | chr10 | 34806087 | - | 6021 | 833 | 20 | 4411 | 1463 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000545693 | PARD3 | chr10 | 34806087 | - | 6243 | 833 | 20 | 4633 | 1537 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000350537 | PARD3 | chr10 | 34806087 | - | 6152 | 833 | 20 | 4543 | 1507 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000346874 | PARD3 | chr10 | 34806087 | - | 6179 | 833 | 20 | 4570 | 1516 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000374789 | PARD3 | chr10 | 34806087 | - | 6290 | 833 | 20 | 4681 | 1553 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000374790 | PARD3 | chr10 | 34806087 | - | 6110 | 833 | 20 | 4501 | 1493 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000374794 | PARD3 | chr10 | 34806087 | - | 5954 | 833 | 20 | 4345 | 1441 |
| ENST00000537177 | CUL2 | chr10 | 35333494 | - | ENST00000374788 | PARD3 | chr10 | 34806087 | - | 6281 | 833 | 20 | 4672 | 1550 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000374746 | ENST00000545260 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000373034 | 0.999627 |
| ENST00000374746 | ENST00000545693 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.00047257 | 0.99952745 |
| ENST00000374746 | ENST00000350537 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000371525 | 0.9996284 |
| ENST00000374746 | ENST00000346874 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000282409 | 0.99971753 |
| ENST00000374746 | ENST00000374789 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000416327 | 0.99958366 |
| ENST00000374746 | ENST00000374790 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000476809 | 0.99952316 |
| ENST00000374746 | ENST00000374794 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000259602 | 0.9997404 |
| ENST00000374746 | ENST00000374788 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000622223 | 0.9993777 |
| ENST00000537177 | ENST00000545260 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000373768 | 0.99962616 |
| ENST00000537177 | ENST00000545693 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000479785 | 0.9995202 |
| ENST00000537177 | ENST00000350537 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000375837 | 0.99962413 |
| ENST00000537177 | ENST00000346874 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000289564 | 0.9997104 |
| ENST00000537177 | ENST00000374789 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000424684 | 0.9995753 |
| ENST00000537177 | ENST00000374790 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000481941 | 0.99951804 |
| ENST00000537177 | ENST00000374794 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000263068 | 0.99973696 |
| ENST00000537177 | ENST00000374788 | CUL2 | chr10 | 35333494 | - | PARD3 | chr10 | 34806087 | - | 0.000634161 | 0.99936587 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >20659_20659_1_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000346874_length(amino acids)=1483AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKD ENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGI HVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNN YYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKK GTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPREL NAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVN GESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALS RIMGESGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQ SMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRII RGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKE KDKMKAKKGMLKGLGDMFRIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKP RNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSS LPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSY -------------------------------------------------------------- >20659_20659_2_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000350537_length(amino acids)=1474AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKD ENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGI HVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNN YYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKK GTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPREL KAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAM ETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVN MPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQIADETKLNTVDDQKAG SPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRES VSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEE RIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVD SNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNA SSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYA -------------------------------------------------------------- >20659_20659_3_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000374788_length(amino acids)=1517AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKD ENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGI HVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNN YYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKK GTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPREL NAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVN GESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALS RIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMS EKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGR GCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDK MKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYG GVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPA TQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARV -------------------------------------------------------------- >20659_20659_4_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000374789_length(amino acids)=1520AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKD ENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGI HVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNN YYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKK GTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPREL NAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVN GESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALS RIMGESGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQ SMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRII RGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKE KDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDEL MYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNA RPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFN -------------------------------------------------------------- >20659_20659_5_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000374790_length(amino acids)=1460AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDE DGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDG DLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSR LPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDR LIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWA KAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKK SSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLER QMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAK TREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQR LRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSP GEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVPPSP -------------------------------------------------------------- >20659_20659_6_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000374794_length(amino acids)=1408AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDE DGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDG DLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSR LPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDR LIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWA KAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGSSPSRDVGPSLGLKKSSSLESLQTAVAEVT LNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRK KDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPR EGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQ EERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQM -------------------------------------------------------------- >20659_20659_7_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000545260_length(amino acids)=1430AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDE DGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDG DLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSR LPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDR LIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWA KAAISDSADCSLSPDVDPVLAFQREGFGRQIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRII RGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKE KDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDEL MYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNA RPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFN -------------------------------------------------------------- >20659_20659_8_CUL2-PARD3_CUL2_chr10_35333494_ENST00000374746_PARD3_chr10_34806087_ENST00000545693_length(amino acids)=1504AA_BP=0 MSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERLYTETKIFLENHVRHLHKRVLESEEQVLVMYHRY WEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMWRKLMVEPLQAILIRMLLREIKNDRGGEDPNQKV IHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYMEKLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGS ELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSEEPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKD ENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGI HVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNN YYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKK GTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPREL KAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAM ETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVN MPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQL DFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKS YDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGD MFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNA RPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQ MQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQR -------------------------------------------------------------- >20659_20659_9_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000346874_length(amino acids)=1516AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKDENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPV GHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHM FRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPS APASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKS QEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELNAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGESGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAG TWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLG LKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHS LERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGG VSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPAT QSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVM -------------------------------------------------------------- >20659_20659_10_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000350537_length(amino acids)=1507AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKDENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPV GHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHM FRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPS APASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKS QEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIIN GGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISH SLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLS PDVDPVLAFQREGFGRQIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAI DKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKG LGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMA LNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSV EVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQ -------------------------------------------------------------- >20659_20659_11_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000374788_length(amino acids)=1550AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKDENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPV GHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHM FRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPS APASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKS QEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELNAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWA KAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKK SSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLER QMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAK TREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQR LRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSP GEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVPPSP -------------------------------------------------------------- >20659_20659_12_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000374789_length(amino acids)=1553AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKDENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPV GHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHM FRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPS APASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKS QEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELNAEPSQMQIPKETKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKEN HADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPI ETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGESGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAG TWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLG LKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHS LERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERI QAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDR IQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQN YSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVP -------------------------------------------------------------- >20659_20659_13_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000374790_length(amino acids)=1493AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGR TLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQ YIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITS RDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTP DGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEG NKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIED DRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSM DLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDE GMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDD KIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMM DALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERES SQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPP -------------------------------------------------------------- >20659_20659_14_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000374794_length(amino acids)=1441AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGR TLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQ YIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITS RDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTP DGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEG NKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIED DRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSM DLGSSPSRDVGPSLGLKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRS GRESVSTASDQPSHSLERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRIQAKTREFRERQARERDYAEIQD FHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDEDVEDRRR TYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRYSSYQGS RNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVPPSPSQVARLNRLQTPEKGRPFY -------------------------------------------------------------- >20659_20659_15_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000545260_length(amino acids)=1463AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKDEDGTEEDNSRVEPVGHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGR TLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHMFRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQ YIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPSAPASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITS RDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKSQEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTP DGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIINGGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEG NKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISHSLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIED DRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLSPDVDPVLAFQREGFGRQIADETKLNTVDDQKAGSPSRDVGPSLG LKKSSSLESLQTAVAEVTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHS LERQMNGNQEKGDKTDRKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERI QAKTREFRERQARERDYAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDR IQRLRQEFQQAKQDEDVEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQN YSPGEGFQSAKENPRYSSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVP -------------------------------------------------------------- >20659_20659_16_CUL2-PARD3_CUL2_chr10_35333494_ENST00000537177_PARD3_chr10_34806087_ENST00000545693_length(amino acids)=1537AA_BP=1 MNAVCRRKQQTLYFMYRVTWSTFWLRFQHYTCTMSLKPRVVDFDETWNKLLTTIKAVVMLEYVERATWNDRFSDIYALCVAYPEPLGERL YTETKIFLENHVRHLHKRVLESEEQVLVMYHRYWEEYSKGADYMDCLYRYLNTQFIKKNKLTEADLQYGYGGVDMNEPLMEIGELALDMW RKLMVEPLQAILIRMLLREIKNDRGGEDPNQKVIHGVINSFVHVEQYKKKFPLKFYQEIFESPFLTETGEYYKQEASNLLQESNCSQYME KLVAVFDEQDPHHGGDGTSASSTGTQSPEIFGSELGTNNVSAFQPYQATSEIEVTPSVLRANMPLHVRRSSDPALIGLSTSVSDSNFSSE EPSRKNPTRWSTTAGFLKQNTAGSPKTCDRKKDENYRSLPRDTSNWSNQFQRDNARSSLSASHPMVGKWLEKQEQDEDGTEEDNSRVEPV GHADTGLEHIPNFSLDDMVKLVEVPNDGGPLGIHVVPFSARGGRTLGLLVKRLEKGGKAEHENLFRENDCIVRINDGDLRNRRFEQAQHM FRQAMRTPIIWFHVVPAANKEQYEQLSQSEKNNYYSSRFSPDSQYIDNRSVNSAGLHTVQRAPRLNHPPEQIDSHSRLPHSAHPSGKPPS APASAPQNVFSTTVSSGYNTKKIGKRLNIQLKKGTEGLGFSITSRDVTIGGSAPIYVKNILPRGAAIQDGRLKAGDRLIEVNGVDLVGKS QEEVVSLLRSTKMEGTVSLLVFRQEDAFHPRELKAEDEDIVLTPDGTREFLTFEVPLNDSGSAGLGVSVKGNRSKENHADLGIFVKSIIN GGAASKDGRLRVNDQLIAVNGESLLGKTNQDAMETLRRSMSTEGNKRGMIQLIVARRISKCNELKSPGSPPGPELPIETALDDRERRISH SLYSGIEGLDESPSRNAALSRIMGKYQLSPTVNMPQDDTVIIEDDRLPVLPPHLSDQSSSSSHDDVGFVTADAGTWAKAAISDSADCSLS PDVDPVLAFQREGFGRQSMSEKRTKQFSDASQLDFVKTRKSKSMDLGIADETKLNTVDDQKAGSPSRDVGPSLGLKKSSSLESLQTAVAE VTLNGDIPFHRPRPRIIRGRGCNESFRAAIDKSYDKPAVDDDDEGMETLEEDTEESSRSGRESVSTASDQPSHSLERQMNGNQEKGDKTD RKKDKTGKEKKKDRDKEKDKMKAKKGMLKGLGDMFRFGKHRKDDKIEKTGKIKIQESFTSEEERIRMKQEQERIQAKTREFRERQARERD YAEIQDFHRTFGCDDELMYGGVSSYEGSMALNARPQSPREGHMMDALYAQVKKPRNSKPSPVDSNRSTPSNHDRIQRLRQEFQQAKQDED VEDRRRTYSFEQPWPNARPATQSGRHSVSVEVQMQRQRQEERESSQQAQRQYSSLPRQSRKNASSVSQDSWEQNYSPGEGFQSAKENPRY SSYQGSRNGYLGGHGFNARVMLETQELLRQEQRRKEQQMKKQPPSEGPSNYDSYKKVQDPSYAPPKGPFRQDVPPSPSQVARLNRLQTPE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:35333494/chr10:34806087) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CUL2 | . |
| FUNCTION: Core component of multiple cullin-RING-based ECS (ElonginB/C-CUL2/5-SOCS-box protein) E3 ubiquitin-protein ligase complexes, which mediate the ubiquitination of target proteins. ECS complexes and ARIH1 collaborate in tandem to mediate ubiquitination of target proteins (PubMed:27565346). May serve as a rigid scaffold in the complex and may contribute to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme. The E3 ubiquitin-protein ligase activity of the complex is dependent on the neddylation of the cullin subunit and is inhibited by the association of the deneddylated cullin subunit with TIP120A/CAND1. The functional specificity of the ECS complex depends on the substrate recognition component. ECS(VHL) mediates the ubiquitination of hypoxia-inducible factor (HIF). {ECO:0000269|PubMed:10973499, ECO:0000269|PubMed:11384984, ECO:0000269|PubMed:27565346, ECO:0000269|PubMed:9122164}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 1049_1077 | 74.0 | 1032.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 1151_1174 | 74.0 | 1032.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 1201_1224 | 74.0 | 1032.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 1280_1301 | 74.0 | 1032.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 1049_1077 | 74.0 | 1320.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 1151_1174 | 74.0 | 1320.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 1201_1224 | 74.0 | 1320.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 1280_1301 | 74.0 | 1320.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 1049_1077 | 74.0 | 1311.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 1151_1174 | 74.0 | 1311.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 1201_1224 | 74.0 | 1311.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 1280_1301 | 74.0 | 1311.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 1049_1077 | 74.0 | 989.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 1151_1174 | 74.0 | 989.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 1201_1224 | 74.0 | 989.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 1280_1301 | 74.0 | 989.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 1049_1077 | 74.0 | 1354.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 1151_1174 | 74.0 | 1354.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 1201_1224 | 74.0 | 1354.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 1280_1301 | 74.0 | 1354.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 1049_1077 | 74.0 | 1357.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 1151_1174 | 74.0 | 1357.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 1201_1224 | 74.0 | 1357.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 1280_1301 | 74.0 | 1357.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 1049_1077 | 74.0 | 1245.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 1151_1174 | 74.0 | 1245.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 1201_1224 | 74.0 | 1245.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 1280_1301 | 74.0 | 1245.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 1049_1077 | 74.0 | 1267.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 1151_1174 | 74.0 | 1267.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 1201_1224 | 74.0 | 1267.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 1280_1301 | 74.0 | 1267.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 1049_1077 | 74.0 | 1341.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 1151_1174 | 74.0 | 1341.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 1201_1224 | 74.0 | 1341.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 1280_1301 | 74.0 | 1341.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 984_1042 | 74.0 | 1032.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 984_1042 | 74.0 | 1320.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 984_1042 | 74.0 | 1311.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 984_1042 | 74.0 | 989.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 984_1042 | 74.0 | 1354.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 984_1042 | 74.0 | 1357.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 984_1042 | 74.0 | 1245.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 984_1042 | 74.0 | 1267.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 984_1042 | 74.0 | 1341.0 | Compositional bias | Note=Lys-rich | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 271_359 | 74.0 | 1032.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 461_546 | 74.0 | 1032.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000340077 | 1 | 21 | 590_677 | 74.0 | 1032.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 271_359 | 74.0 | 1320.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 461_546 | 74.0 | 1320.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000346874 | 1 | 24 | 590_677 | 74.0 | 1320.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 271_359 | 74.0 | 1311.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 461_546 | 74.0 | 1311.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000350537 | 1 | 24 | 590_677 | 74.0 | 1311.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 271_359 | 74.0 | 989.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 461_546 | 74.0 | 989.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374776 | 1 | 20 | 590_677 | 74.0 | 989.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 271_359 | 74.0 | 1354.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 461_546 | 74.0 | 1354.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374788 | 1 | 25 | 590_677 | 74.0 | 1354.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 271_359 | 74.0 | 1357.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 461_546 | 74.0 | 1357.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374789 | 1 | 25 | 590_677 | 74.0 | 1357.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 271_359 | 74.0 | 1245.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 461_546 | 74.0 | 1245.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000374794 | 1 | 21 | 590_677 | 74.0 | 1245.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 271_359 | 74.0 | 1267.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 461_546 | 74.0 | 1267.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545260 | 1 | 23 | 590_677 | 74.0 | 1267.0 | Domain | PDZ 3 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 271_359 | 74.0 | 1341.0 | Domain | PDZ 1 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 461_546 | 74.0 | 1341.0 | Domain | PDZ 2 | |
| Tgene | PARD3 | chr10:35333494 | chr10:34806087 | ENST00000545693 | 1 | 24 | 590_677 | 74.0 | 1341.0 | Domain | PDZ 3 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CUL2 | |
| PARD3 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CUL2-PARD3 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CUL2-PARD3 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

