| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ETV6-ABL2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ETV6-ABL2 | FusionPDB ID: 27698 | FusionGDB2.0 ID: 27698 | Hgene | Tgene | Gene symbol | ETV6 | ABL2 | Gene ID | 2120 | 27 |
| Gene name | ETS variant transcription factor 6 | ABL proto-oncogene 2, non-receptor tyrosine kinase | |
| Synonyms | TEL|TEL/ABL|THC5 | ABLL|ARG | |
| Cytomap | 12p13.2 | 1q25.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcription factor ETV6ETS translocation variant 6ETS variant 6ETS-related protein Tel1TEL1 oncogeneets variant gene 6 (TEL oncogene) | tyrosine-protein kinase ABL2Abelson tyrosine-protein kinase 2abelson-related gene proteinc-abl oncogene 2, non-receptor tyrosine kinasetyrosine-protein kinase ARGv-abl Abelson murine leukemia viral oncogene homolog 2 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | P41212 | P42684 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000396373, ENST00000544715, | ENST00000344730, ENST00000367623, ENST00000392043, ENST00000408940, ENST00000504405, ENST00000507173, ENST00000511413, ENST00000512653, ENST00000502732, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 59 X 37 X 25=54575 | 12 X 13 X 3=468 |
| # samples | 58 | 13 | |
| ** MAII score | log2(58/54575*10)=-6.55604351475058 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/468*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ETV6 [Title/Abstract] AND ABL2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ETV6 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 10514502 |
| Tgene | ABL2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 15886098 |
| Tgene | ABL2 | GO:0051353 | positive regulation of oxidoreductase activity | 12893824 |
 Fusion gene breakpoints across ETV6 (5'-gene) Fusion gene breakpoints across ETV6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across ABL2 (3'-gene) Fusion gene breakpoints across ABL2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | ETV6 | chr12 | 12022903 | + | ABL2 | chr1 | 179102509 | - |
| ChimerKB4 | . | . | ETV6 | chr12 | 12022903 | + | ABL2 | chr1 | 179102509 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396373 | ETV6 | chr12 | 12022903 | + | ENST00000502732 | ABL2 | chr1 | 179102509 | - | 13066 | 1283 | 274 | 4674 | 1466 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >27698_27698_1_ETV6-ABL2_ETV6_chr12_12022903_ENST00000396373_ABL2_chr1_179102509_ENST00000502732_length(amino acids)=1466AA_BP=336 MSETPAQCSIKQERISYTPPESPVPSYASSTPLHVPVPRALRMEEDSIRLPAHLRLQPIYWSRDDVAQWLKWAENEFSLRPIDSNTFEMN GKALLLLTKEDFRYRSPHSGDVLYELLQHILKQRKPRILFSPFFHPGNSIHTQPEVILHQNHEEDNCVQRTPRPSVDNVHHNPPTIELLH RSRSPITTNHRPSPDPEQRPLRSPLDNMIRRLSPAERAQGPRPHQENNHQESYPLSVSPMENNHCPASSESHPKPSSPRQESTRVIQLMP SPIMHPLILNPRHSVDFKQSRLSEDGLHREGKPINLSHREDLAYMNHIMVSVSPPEEHAMPIGRIADHFASCVEDGFEGDKTGGSSPEAL HRPYGCDVEPQALNEAIRWSSKENLLGATESDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNQNGEWSEVRSKNGQGWVPSNYITPVN SLEKHSWYHGPVSRSAAEYLLSSLINGSFLVRESESSPGQLSISLRYEGRVYHYRINTTADGKVYVTAESRFSTLAELVHHHSTVADGLV TTLHYPAPKCNKPTVYGVSPIHDKWEMERTDITMKHKLGGGQYGEVYVGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQ LLGVCTLEPPFYIVTEYMPYGNLLDYLRECNREEVTAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHVVKVADFGLSRLMTG DTYTAHAGAKFPIKWTAPESLAYNTFSIKSDVWAFGVLLWEIATYGMSPYPGIDLSQVYDLLEKGYRMEQPEGCPPKVYELMRACWKWSP ADRPSFAETHQAFETMFHDSSISEEVAEELGRAASSSSVVPYLPRLPILPSKTRTLKKQVENKENIEGAQDATENSASSLAPGFIRGAQA SSGSPALPRKQRDKSPSSLLEDAKETCFTRDRKGGFFSSFMKKRNAPTPPKRSSSFREMENQPHKKYELTGNFSSVASLQHADGFSFTPA QQEANLVPPKCYGGSFAQRNLCNDDGGGGGGSGTAGGGWSGITGFFTPRLIKKTLGLRAGKPTASDDTSKPFPRSNSTSSMSSGLPEQDR MAMTLPRNCQRSKLQLERTVSTSSQPEENVDRANDMLPKKSEESAAPSRERPKAKLLPRGATALPLRTPSGDLAITEKDPPGVGVAGVAA APKGKEKNGGARLGMAGVPEDGEQPGWPSPAKAAPVLPTTHNHKVPVLISPTLKHTPADVQLIGTDSQGNKFKLLSEHQVTSSGDKDRPR RVKPKCAPPPPPVMRLLQHPSICSDPTEEPTALTAGQSTSETQEGGKKAALGAVPISGKAGRPVMPPPQVPLPTSSISPAKMANGTAGTK VALRKTKQAAEKISADKISKEALLECADLLSSALTEPVPNSQLVDTGHQLLDYCSGYVDCIPQTRNKFAFREAVSKLELSLQELQVSSAA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:/chr1:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ETV6 | ABL2 |
| FUNCTION: Transcriptional repressor; binds to the DNA sequence 5'-CCGGAAGT-3'. Plays a role in hematopoiesis and malignant transformation. {ECO:0000269|PubMed:25581430}. | FUNCTION: Non-receptor tyrosine-protein kinase that plays an ABL1-overlapping role in key processes linked to cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion and receptor endocytosis. Coordinates actin remodeling through tyrosine phosphorylation of proteins controlling cytoskeleton dynamics like MYH10 (involved in movement); CTTN (involved in signaling); or TUBA1 and TUBB (microtubule subunits). Binds directly F-actin and regulates actin cytoskeletal structure through its F-actin-bundling activity. Involved in the regulation of cell adhesion and motility through phosphorylation of key regulators of these processes such as CRK, CRKL, DOK1 or ARHGAP35. Adhesion-dependent phosphorylation of ARHGAP35 promotes its association with RASA1, resulting in recruitment of ARHGAP35 to the cell periphery where it inhibits RHO. Phosphorylates multiple receptor tyrosine kinases like PDGFRB and other substrates which are involved in endocytosis regulation such as RIN1. In brain, may regulate neurotransmission by phosphorylating proteins at the synapse. ABL2 acts also as a regulator of multiple pathological signaling cascades during infection. Pathogens can highjack ABL2 kinase signaling to reorganize the host actin cytoskeleton for multiple purposes, like facilitating intracellular movement and host cell exit. Finally, functions as its own regulator through autocatalytic activity as well as through phosphorylation of its inhibitor, ABI1. {ECO:0000269|PubMed:15735735, ECO:0000269|PubMed:15886098, ECO:0000269|PubMed:16678104, ECO:0000269|PubMed:17306540, ECO:0000269|PubMed:18945674}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (935)CIF file (935) >>>935.cif | ETV6 | chr12 | 12022903 | + | ABL2 | chr1 | 179102509 | - | MSETPAQCSIKQERISYTPPESPVPSYASSTPLHVPVPRALRMEEDSIRL PAHLRLQPIYWSRDDVAQWLKWAENEFSLRPIDSNTFEMNGKALLLLTKE DFRYRSPHSGDVLYELLQHILKQRKPRILFSPFFHPGNSIHTQPEVILHQ NHEEDNCVQRTPRPSVDNVHHNPPTIELLHRSRSPITTNHRPSPDPEQRP LRSPLDNMIRRLSPAERAQGPRPHQENNHQESYPLSVSPMENNHCPASSE SHPKPSSPRQESTRVIQLMPSPIMHPLILNPRHSVDFKQSRLSEDGLHRE GKPINLSHREDLAYMNHIMVSVSPPEEHAMPIGRIADHFASCVEDGFEGD KTGGSSPEALHRPYGCDVEPQALNEAIRWSSKENLLGATESDPNLFVALY DFVASGDNTLSITKGEKLRVLGYNQNGEWSEVRSKNGQGWVPSNYITPVN SLEKHSWYHGPVSRSAAEYLLSSLINGSFLVRESESSPGQLSISLRYEGR VYHYRINTTADGKVYVTAESRFSTLAELVHHHSTVADGLVTTLHYPAPKC NKPTVYGVSPIHDKWEMERTDITMKHKLGGGQYGEVYVGVWKKYSLTVAV KTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCTLEPPFYIVTEYMPY GNLLDYLRECNREEVTAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCL VGENHVVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNTFSIKS DVWAFGVLLWEIATYGMSPYPGIDLSQVYDLLEKGYRMEQPEGCPPKVYE LMRACWKWSPADRPSFAETHQAFETMFHDSSISEEVAEELGRAASSSSVV PYLPRLPILPSKTRTLKKQVENKENIEGAQDATENSASSLAPGFIRGAQA SSGSPALPRKQRDKSPSSLLEDAKETCFTRDRKGGFFSSFMKKRNAPTPP KRSSSFREMENQPHKKYELTGNFSSVASLQHADGFSFTPAQQEANLVPPK CYGGSFAQRNLCNDDGGGGGGSGTAGGGWSGITGFFTPRLIKKTLGLRAG KPTASDDTSKPFPRSNSTSSMSSGLPEQDRMAMTLPRNCQRSKLQLERTV STSSQPEENVDRANDMLPKKSEESAAPSRERPKAKLLPRGATALPLRTPS GDLAITEKDPPGVGVAGVAAAPKGKEKNGGARLGMAGVPEDGEQPGWPSP AKAAPVLPTTHNHKVPVLISPTLKHTPADVQLIGTDSQGNKFKLLSEHQV TSSGDKDRPRRVKPKCAPPPPPVMRLLQHPSICSDPTEEPTALTAGQSTS ETQEGGKKAALGAVPISGKAGRPVMPPPQVPLPTSSISPAKMANGTAGTK VALRKTKQAAEKISADKISKEALLECADLLSSALTEPVPNSQLVDTGHQL LDYCSGYVDCIPQTRNKFAFREAVSKLELSLQELQVSSAAAGVPGTNPVL | 1466 |
| 3D view using mol* of 935 (AA BP:) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ETV6_pLDDT.png |
ABL2_pLDDT.png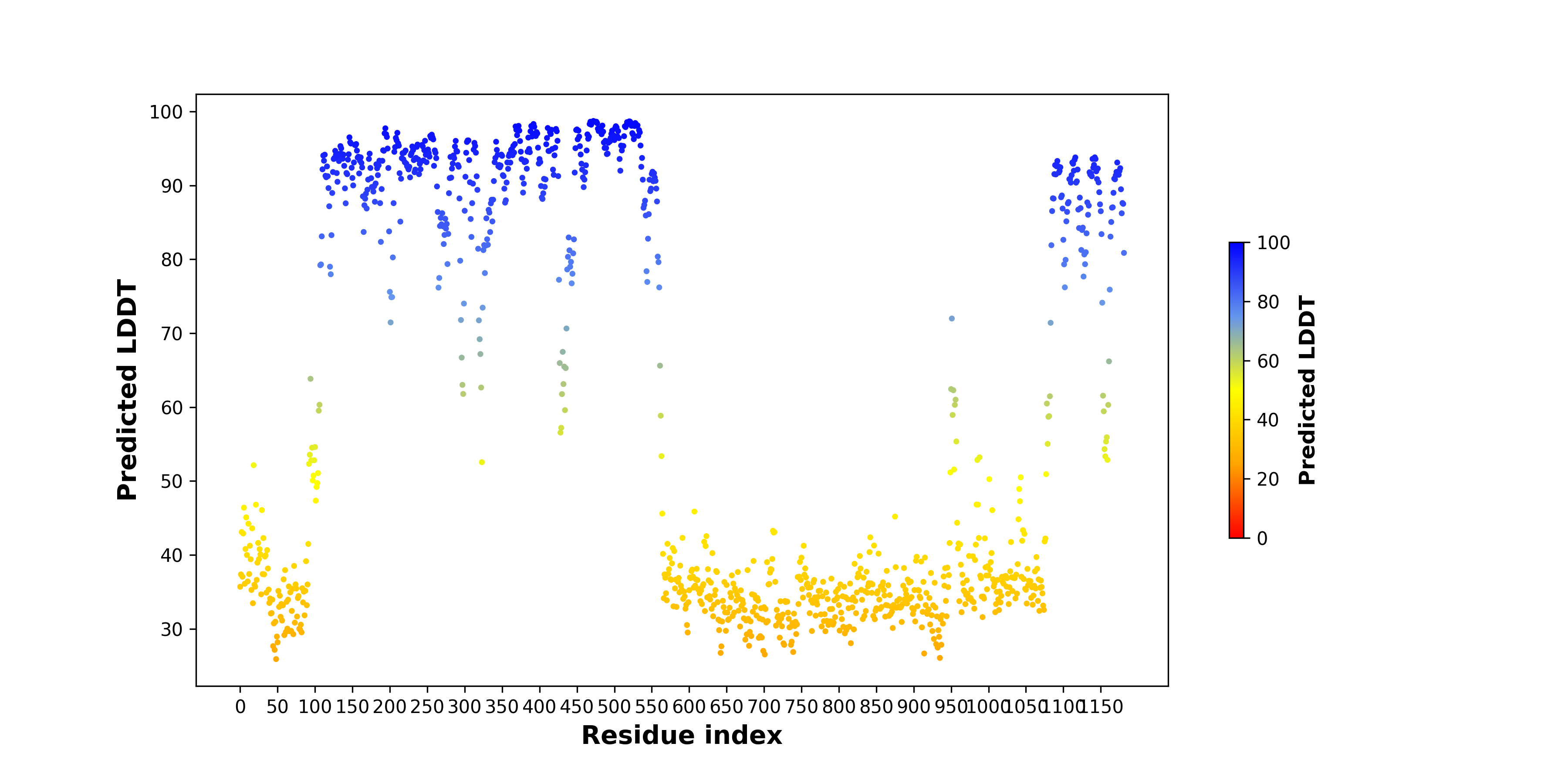 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ETV6_ABL2_935_pLDDT_and_active_sites.png (AA BP:)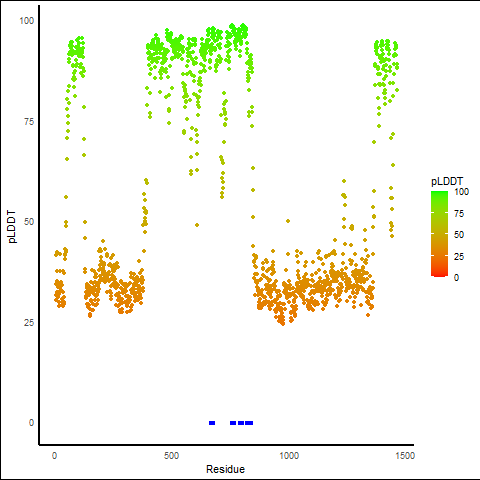 |
ETV6_ABL2_935_violinplot.png (AA BP:)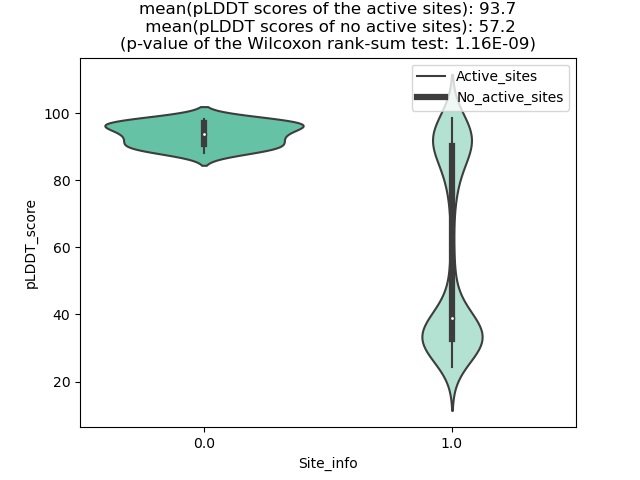 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 935 | 1.081 | 97 | 1.125 | 194.481 | 0.492 | 0.767 | 1.071 | 2.894 | 0.799 | 3.622 | 0.995 | Chain A: 662,666,667,670,671,674,759,762,763,764,7 65,767,791,792,793,794,795,798,823,832,833,836,840 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 935 | ZINC000005844788 | DB04861 | Nebivolol | -9.23094 | -9.25074 |
| 935 | ZINC000000000850 | DB00744 | Zileuton | -8.98931 | -9.00791 |
| 935 | ZINC000043206370 | DB11793 | Niraparib | -8.85208 | -8.85208 |
| 935 | ZINC000004213946 | DB04861 | Nebivolol | -8.64132 | -8.66112 |
| 935 | ZINC000035342789 | DB00323 | Tolcapone | -7.06468 | -8.60168 |
| 935 | ZINC000001999441 | DB04861 | Nebivolol | -8.42557 | -8.44537 |
| 935 | ZINC000053084692 | DB06654 | Safinamide | -7.65969 | -8.43779 |
| 935 | ZINC000005844792 | DB04861 | Nebivolol | -8.40519 | -8.42499 |
| 935 | Infigratinib | DB04861 | -7.85022 | -8.41702 | |
| 935 | ZINC000043207238 | DB08907 | Canagliflozin | -8.31459 | -8.31459 |
| 935 | ZINC000034220093 | DB15058 | Flutemetamol | -8.24466 | -8.25876 |
| 935 | ZINC000000000323 | DB00712 | Flurbiprofen | -8.25155 | -8.25205 |
| 935 | ZINC000000008667 | DB00712 | Flurbiprofen | -8.22911 | -8.22961 |
| 935 | ZINC000006467621 | DB11115 | Ensulizole | -7.80972 | -8.22062 |
| 935 | ZINC000000643143 | DB01026 | Ketoconazole | -7.80084 | -8.12554 |
| 935 | ZINC000000020243 | DB00861 | Diflunisal | -8.06762 | -8.06762 |
| 935 | ZINC000000004319 | DB00598 | Labetalol | -8.02603 | -8.05363 |
| 935 | ZINC000003872605 | DB00176 | Fluvoxamine | -8.04084 | -8.04444 |
| 935 | ZINC000252678020 | DB00176 | Fluvoxamine | -8.04084 | -8.04444 |
| 935 | ZINC000002568036 | DB01219 | Dantrolene | -7.92536 | -7.99776 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000043206370 | DB11793 | Niraparib | Small molecule | NC(=O)C1=CC=CC2=CN(N=C12)C1=CC=C(C=C1)[C@@H]1CCCNC1 | Approved|Investigational |
| ZINC000035342789 | DB00323 | Tolcapone | Small molecule | CC1=CC=C(C=C1)C(=O)C1=CC(=C(O)C(O)=C1)[N+]([O-])=O | Approved|Withdrawn |
| ZINC000053084692 | DB06654 | Safinamide | Small molecule | C[C@H](NCC1=CC=C(OCC2=CC(F)=CC=C2)C=C1)C(N)=O | Approved|Investigational |
| ZINC000043207238 | DB08907 | Canagliflozin | Small molecule | [H][C@]1(O[C@H](CO)[C@@H](O)[C@H](O)[C@H]1O)C1=CC=C(C)C(CC2=CC=C(S2)C2=CC=C(F)C=C2)=C1 | Approved |
| ZINC000034220093 | DB15058 | Flutemetamol | Small molecule | CNC1=C(F)C=C(C=C1)C1=NC2=C(S1)C=C(O)C=C2 | Investigational |
| ZINC000000008667 | DB00712 | Flurbiprofen | Small molecule | CC(C(O)=O)C1=CC(F)=C(C=C1)C1=CC=CC=C1 | Approved|Investigational |
| ZINC000006467621 | DB11115 | Ensulizole | Small molecule | OS(=O)(=O)C1=CC2=C(C=C1)N=C(N2)C1=CC=CC=C1 | Approved |
| ZINC000000020243 | DB00861 | Diflunisal | Small molecule | OC(=O)C1=C(O)C=CC(=C1)C1=C(F)C=C(F)C=C1 | Approved|Investigational |
| ZINC000252678020 | DB00176 | Fluvoxamine | Small molecule | COCCCCC(=N/OCCN)C1=CC=C(C=C1)C(F)(F)F | Approved|Investigational |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000005844788 | 405.441 | 1.102 | 724.6 | 273.703 | 87.795 | 269.339 | 93.762 | 1272.257 | 3 | 6.4 | 9.305 | 3 | 94.995 | 0 | 0 |
| ZINC000005844788 | 405.441 | 3.204 | 688.14 | 293.004 | 60.761 | 240.615 | 93.76 | 1244.959 | 3 | 6.4 | 9.151 | 3 | 100 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 457.642 | 85.783 | 137.035 | 198.523 | 36.3 | 755.015 | 3 | 3.7 | 8.91 | 3 | 75.351 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 450.837 | 85.532 | 135.419 | 201.893 | 27.994 | 746.839 | 3 | 3.7 | 8.57 | 3 | 76.555 | 0 | 0 |
| ZINC000043206370 | 320.393 | 3.754 | 607.688 | 185.794 | 143.417 | 278.476 | 0 | 1055.805 | 3 | 5 | 8.546 | 3 | 76.72 | 0 | 0 |
| ZINC000004213946 | 405.441 | 5.213 | 712.775 | 270.546 | 87.695 | 260.773 | 93.761 | 1260.01 | 3 | 6.4 | 9.251 | 3 | 94.555 | 0 | 0 |
| ZINC000004213946 | 405.441 | 3.313 | 660.255 | 261.068 | 75.583 | 229.844 | 93.76 | 1216.093 | 3 | 6.4 | 9.124 | 3 | 95.533 | 0 | 0 |
| ZINC000035342789 | 273.245 | 3.817 | 507.142 | 88.195 | 223.7 | 195.247 | 0 | 847.548 | 2 | 4.5 | 9.508 | 3 | 67.934 | 0 | 0 |
| ZINC000035342789 | 273.245 | 5.603 | 506.169 | 88.195 | 222.204 | 195.769 | 0 | 846.891 | 2 | 4.5 | 9.828 | 3 | 68.233 | 0 | 0 |
| ZINC000035342789 | 273.245 | 3.5 | 506.842 | 88.2 | 223.976 | 194.665 | 0 | 847.734 | 2 | 4.5 | 9.758 | 3 | 67.881 | 0 | 0 |
| ZINC000001999441 | 405.441 | 4.722 | 712.226 | 273.488 | 84.211 | 260.774 | 93.753 | 1256.147 | 3 | 6.4 | 9.229 | 3 | 95.143 | 0 | 0 |
| ZINC000001999441 | 405.441 | 0.513 | 723.645 | 269.655 | 91.189 | 269.032 | 93.768 | 1265.844 | 3 | 6.4 | 9.27 | 3 | 94.039 | 0 | 0 |
| ZINC000053084692 | 302.348 | 6.36 | 612.581 | 156.033 | 105.388 | 304.286 | 46.874 | 1036.218 | 3 | 4.75 | 9.083 | 3 | 76.961 | 0 | 0 |
| ZINC000053084692 | 302.348 | 3.053 | 603.233 | 143.432 | 120.534 | 292.37 | 46.897 | 1022.597 | 3 | 4.75 | 9.137 | 3 | 72.683 | 0 | 0 |
| ZINC000005844792 | 405.441 | 2.88 | 705.095 | 278.739 | 76.814 | 255.781 | 93.761 | 1255.099 | 3 | 6.4 | 9.355 | 3 | 96.665 | 0 | 0 |
| ZINC000005844792 | 405.441 | 2.543 | 700.215 | 280.743 | 73.672 | 252.036 | 93.764 | 1255.101 | 3 | 6.4 | 9.361 | 3 | 100 | 0 | 0 |
| ZINC000043207238 | 444.517 | 4.415 | 747.287 | 209.678 | 166.727 | 291.446 | 79.436 | 1342.618 | 4 | 8.5 | 8.906 | 3 | 88.99 | 0 | 2 |
| ZINC000034220093 | 274.312 | 1.949 | 511.177 | 89.833 | 86.339 | 254.76 | 80.245 | 847.322 | 2 | 3.25 | 8.207 | 3 | 100 | 0 | 0 |
| ZINC000034220093 | 274.312 | 1.174 | 509.609 | 89.83 | 84.419 | 255.176 | 80.184 | 844.623 | 2 | 3.25 | 8.146 | 3 | 100 | 0 | 0 |
| ZINC000000000323 | 244.265 | 2.669 | 490.209 | 87.743 | 92.913 | 277.089 | 32.464 | 818.983 | 1 | 2 | 9.231 | 3 | 96.354 | 0 | 0 |
| ZINC000000008667 | 244.265 | 2.669 | 490.209 | 87.743 | 92.913 | 277.089 | 32.464 | 818.983 | 1 | 2 | 9.231 | 3 | 96.354 | 0 | 0 |
| ZINC000006467621 | 274.294 | 9.514 | 495.179 | 0 | 165.642 | 327.077 | 2.46 | 816.857 | 2 | 5.5 | 9.09 | 3 | 70.241 | 0 | 0 |
| ZINC000006467621 | 274.294 | 5.937 | 496.167 | 0 | 165.586 | 328.124 | 2.456 | 817.805 | 2 | 5.5 | 9.121 | 3 | 70.288 | 0 | 0 |
| ZINC000006467621 | 274.294 | 9.57 | 496.894 | 0 | 166.022 | 328.377 | 2.495 | 819.646 | 2 | 5.5 | 8.954 | 3 | 70.264 | 0 | 0 |
| ZINC000000643143 | 531.438 | 9.855 | 856.851 | 327.713 | 80.224 | 331.811 | 117.103 | 1560.895 | 0 | 8.25 | 8.211 | 3 | 92.471 | 1 | 0 |
| ZINC000000643143 | 531.438 | 6.997 | 824.307 | 333.326 | 81.685 | 289.695 | 119.601 | 1537.566 | 0 | 8.25 | 8.204 | 3 | 92.427 | 1 | 0 |
| ZINC000000643143 | 531.438 | 6.356 | 853.952 | 332.548 | 78.446 | 320.618 | 122.341 | 1564.061 | 0 | 8.25 | 9.22 | 3 | 92.922 | 1 | 0 |
| ZINC000000643143 | 531.438 | 6.331 | 852.73 | 332.662 | 78.89 | 318.374 | 122.804 | 1561.366 | 0 | 8.25 | 9.213 | 3 | 92.807 | 1 | 0 |
| ZINC000000020243 | 250.201 | 8.261 | 459.081 | 0 | 144.436 | 231.005 | 83.64 | 751.879 | 1 | 1.75 | 9.879 | 3 | 86.601 | 0 | 0 |
| ZINC000000004319 | 328.41 | 5.655 | 664.549 | 169.051 | 199.646 | 295.852 | 0 | 1140.615 | 4 | 5.45 | 9.067 | 2 | 70.589 | 0 | 0 |
| ZINC000000004319 | 328.41 | 3.937 | 652.909 | 170.739 | 198.631 | 283.539 | 0 | 1134.882 | 4 | 5.45 | 9.364 | 2 | 70.662 | 0 | 0 |
| ZINC000003872605 | 318.338 | 3.821 | 606.238 | 306.607 | 65.012 | 116.869 | 117.75 | 1044.621 | 2 | 5.4 | 9.599 | 3 | 93.672 | 0 | 0 |
| ZINC000252678020 | 318.338 | 3.821 | 606.238 | 306.607 | 65.012 | 116.869 | 117.75 | 1044.621 | 2 | 5.4 | 9.599 | 3 | 93.672 | 0 | 0 |
| ZINC000252678020 | 318.338 | 3.426 | 598.781 | 317.355 | 53.672 | 110.076 | 117.678 | 1044.796 | 2 | 5.4 | 9.44 | 3 | 96.425 | 0 | 0 |
| ZINC000002568036 | 314.257 | 3.867 | 548.758 | 54.323 | 243.185 | 251.25 | 0 | 924.46 | 1 | 5 | 9.205 | 3 | 67.058 | 0 | 0 |
| ZINC000002568036 | 314.257 | 4.186 | 547.3 | 54.729 | 241.127 | 251.444 | 0 | 922.097 | 1 | 5 | 9.191 | 3 | 67.408 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000005844788 | O[C@@H](CNC[C@@H](O)[C@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000000000850 | C[C@@H](c1cc2ccccc2s1)N(O)C(N)=O | 0.136335882 | 0.223511692 |
| ZINC000043206370 | NC(=O)c1cccc2cn(-c3ccc([C@@H]4CCCNC4)cc3)nc12 | 0.15810306 | 0.243905197 |
| ZINC000004213946 | O[C@@H](CNC[C@@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000035342789 | Cc1ccc(C(=O)c2cc(O)c(O)c([N+](=O)[O-])c2)cc1 | 0.359735014 | 0.498165074 |
| ZINC000001999441 | O[C@@H](CNC[C@@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000053084692 | C[C@H](NCc1ccc(OCc2cccc(F)c2)cc1)C(N)=O | 0.279955479 | 0.265373208 |
| ZINC000005844792 | O[C@@H](CNC[C@H](O)[C@@H]1CCc2cc(F)ccc2O1)[C@@H]1CCc2cc(F)ccc2O1 | 0.055550082 | 0.198725964 |
| ZINC000043207238 | Cc1ccc([C@@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)cc1Cc1ccc(-c2ccc(F)cc2)s1 | 0.068211161 | 0.161061761 |
| ZINC000034220093 | CNc1ccc(-c2nc3ccc(O)cc3s2)cc1F | 0.301158621 | 0.239116152 |
| ZINC000000000323 | C[C@H](C(=O)O)c1ccc(-c2ccccc2)c(F)c1 | 0.31380464 | 0.287418039 |
| ZINC000000008667 | C[C@@H](C(=O)O)c1ccc(-c2ccccc2)c(F)c1 | 0.31380464 | 0.287418039 |
| ZINC000006467621 | O=S(=O)(O)c1ccc2[nH]c(-c3ccccc3)nc2c1 | 0.38504104 | 0.405031793 |
| ZINC000000643143 | CC(=O)N1CCN(c2ccc(OC[C@@H]3CO[C@](Cn4ccnc4)(c4ccc(Cl)cc4Cl)O3)cc2)CC1 | 0.087393272 | 0.387082142 |
| ZINC000000020243 | O=C(O)c1cc(-c2ccc(F)cc2F)ccc1O | 0.440409373 | 0.371648269 |
| ZINC000000004319 | C[C@H](CCc1ccccc1)NC[C@H](O)c1ccc(O)c(C(N)=O)c1 | 0.163888793 | 0.349082082 |
| ZINC000003872605 | COCCCC/C(=NOCCN)c1ccc(C(F)(F)F)cc1 | 0.232252376 | 0.356848479 |
| ZINC000252678020 | COCCCCC(=NOCCN)c1ccc(C(F)(F)F)cc1 | 0.232252376 | 0.356848479 |
| ZINC000002568036 | O=C1CN(/N=C/c2ccc(-c3ccc([N+](=O)[O-])cc3)o2)C(=O)N1 | 0.204070823 | 0.461176527 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ETV6 | KAT5, GAB2, ETV6, GRB2, CRKL, ACTA1, HDAC9, FLI1, FBXL6, SKP1, L3MBTL1, IRF8, NCOR1, SIN3A, HDAC3, UBE2I, ELAVL1, PDGFRB, SUMO1, SOCS1, SOCS3, TRAF3, SOX2, HDAC6, PIN1, AP1M1, WDYHV1, LRP6, LRP5, NID2, ETV7, USP7, VPS26B, NFATC2, EGLN3, NKX2-1, TRAF2, AXIN1, ESR2, LMNA, TP53BP1, BRCA1, MDC1, KIAA1429, WWP2, ESR1, BRD4, NFE2L2, MAD2L1, B4GALT2, KLHL9, KLHL13, SUMO2, ARHGAP32, SEC16A, XPOT, PIAS2, CEP152, NUP214, NUP62, NUP98, SEC13, RBM11, ECH1, ZNF146, TRIP6, CEP85, NUP54, TAB3, ALMS1, CEP192, PPIL4, RQCD1, SDCCAG3, NUP88, TBL1Y, KIAA1671, C2orf44, GRPEL1, TNRC6C, MID1IP1, CYLD, ZMYM2, LIMD1, SPATA2, TNRC6B, TIMM13, FLOT1, LGALS3BP, FLOT2, HNRNPUL1, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ETV6 |  |
| ABL2 | 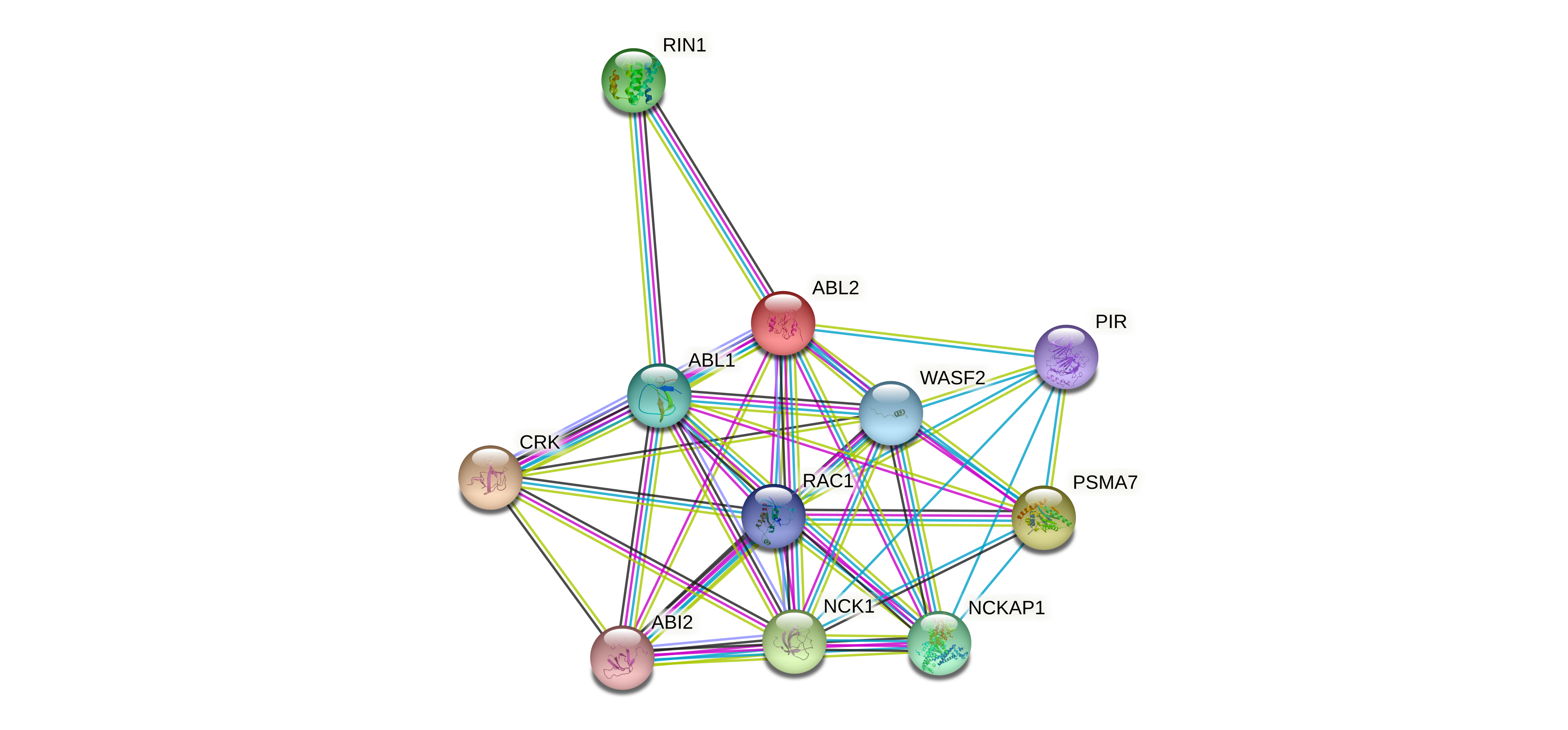 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ETV6-ABL2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ETV6-ABL2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| ETV6 | ABL2 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| ETV6 | ABL2 | Invasive Breast Carcinoma | MyCancerGenome | |
| ETV6 | ABL2 | Lung Adenocarcinoma | MyCancerGenome | |
| ETV6 | ABL2 | Appendix Adenocarcinoma | MyCancerGenome | |
| ETV6 | ABL2 | Colon Adenocarcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ETV6 | C4015537 | THROMBOCYTOPENIA 5 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | ETV6 | C0023485 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma | 3 | CTD_human |
| Hgene | ETV6 | C0040034 | Thrombocytopenia | 3 | CTD_human;GENOMICS_ENGLAND |
| Hgene | ETV6 | C0023480 | Leukemia, Myelomonocytic, Chronic | 2 | ORPHANET |
| Hgene | ETV6 | C1332965 | Congenital Mesoblastic Nephroma | 2 | ORPHANET |
| Hgene | ETV6 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | ETV6 | C0013146 | Drug abuse | 1 | CTD_human |
| Hgene | ETV6 | C0013170 | Drug habituation | 1 | CTD_human |
| Hgene | ETV6 | C0013222 | Drug Use Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ETV6 | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | ETV6 | C0023467 | Leukemia, Myelocytic, Acute | 1 | CGI;CTD_human;GENOMICS_ENGLAND |
| Hgene | ETV6 | C0029231 | Organic Mental Disorders, Substance-Induced | 1 | CTD_human |
| Hgene | ETV6 | C0038580 | Substance Dependence | 1 | CTD_human |
| Hgene | ETV6 | C0038586 | Substance Use Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Hgene | ETV6 | C0236969 | Substance-Related Disorders | 1 | CTD_human |
| Hgene | ETV6 | C0238463 | Papillary thyroid carcinoma | 1 | ORPHANET |
| Hgene | ETV6 | C0376544 | Hematopoietic Neoplasms | 1 | CTD_human |
| Hgene | ETV6 | C0376545 | Hematologic Neoplasms | 1 | CTD_human |
| Hgene | ETV6 | C0740858 | Substance abuse problem | 1 | CTD_human |
| Hgene | ETV6 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Hgene | ETV6 | C1510472 | Drug Dependence | 1 | CTD_human |
| Hgene | ETV6 | C1832388 | Platelet Disorder, Familial, with Associated Myeloid Malignancy | 1 | ORPHANET |
| Hgene | ETV6 | C1838656 | Macrocytosis, Familial | 1 | CTD_human |
| Hgene | ETV6 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | ETV6 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Hgene | ETV6 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Hgene | ETV6 | C4316881 | Prescription Drug Abuse | 1 | CTD_human |
| Hgene | ETV6 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Hgene | ETV6 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |

