| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:AHNAK-BIRC6 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: AHNAK-BIRC6 | FusionPDB ID: 3105 | FusionGDB2.0 ID: 3105 | Hgene | Tgene | Gene symbol | AHNAK | BIRC6 | Gene ID | 79026 | 57448 |
| Gene name | AHNAK nucleoprotein | baculoviral IAP repeat containing 6 | |
| Synonyms | AHNAKRS|PM227 | APOLLON|BRUCE | |
| Cytomap | 11q12.3 | 2p22.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | neuroblast differentiation-associated protein AHNAKAHNAK-relateddesmoyokin | baculoviral IAP repeat-containing protein 6BIR repeat-containing ubiquitin-conjugating enzymeRING-type E3 ubiquitin transferase BIRC6ubiquitin-conjugating BIR-domain enzyme apollon | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q8IVF2 | Q9NR09 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000378024, ENST00000257247, ENST00000525875, ENST00000530124, | ENST00000421745, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 27 X 21 X 12=6804 | 23 X 19 X 14=6118 |
| # samples | 34 | 29 | |
| ** MAII score | log2(34/6804*10)=-4.32277648952568 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(29/6118*10)=-4.39893530154327 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: AHNAK [Title/Abstract] AND BIRC6 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | AHNAK(62283375)-BIRC6(32820101), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AHNAK-BIRC6 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AHNAK-BIRC6 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AHNAK-BIRC6 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AHNAK-BIRC6 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across AHNAK (5'-gene) Fusion gene breakpoints across AHNAK (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
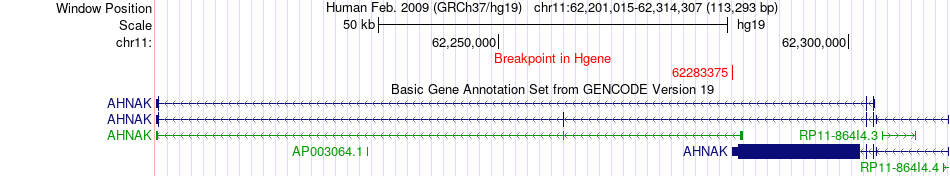 |
 Fusion gene breakpoints across BIRC6 (3'-gene) Fusion gene breakpoints across BIRC6 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
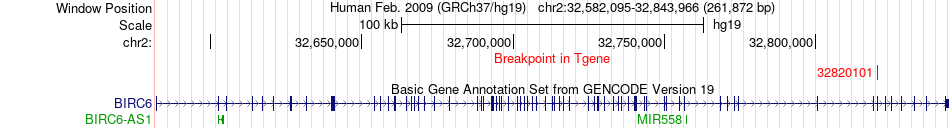 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-EE-A2GT-06A | AHNAK | chr11 | 62283375 | - | BIRC6 | chr2 | 32820101 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000378024 | AHNAK | chr11 | 62283375 | - | ENST00000421745 | BIRC6 | chr2 | 32820101 | + | 20855 | 18787 | 275 | 17947 | 5890 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000378024 | ENST00000421745 | AHNAK | chr11 | 62283375 | - | BIRC6 | chr2 | 32820101 | + | 0.000237308 | 0.9997627 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >3105_3105_1_AHNAK-BIRC6_AHNAK_chr11_62283375_ENST00000378024_BIRC6_chr2_32820101_ENST00000421745_length(amino acids)=5890AA_BP= MEKEETTRELLLPNWQGSGSHGLTIAQRDDGVFVQEVTQNSPAARTGVVKEGDQIVGATIYFDNLQSGEVTQLLNTMGHHTVGLKLHRKG DRSPEPGQTWTREVFSSCSSEVVLSGDDEEYQRIYTTKIKPRLKSEDGVEGDLGETQSRTITVTRRVTAYTVDVTGREGAKDIDISSPEF KIKIPRHELTEISNVDVETQSGKTVIRLPSGSGAASPTGSAVDIRAGAISASGPELQGAGHSKLQVTMPGIKVGGSGVNVNAKGLDLGGR GGVQVPAVDISSSLGGRAVEVQGPSLESGDHGKIKFPTMKVPKFGVSTGREGQTPKAGLRVSAPEVSVGHKGGKPGLTIQAPQLEVSVPS ANIEGLEGKLKGPQITGPSLEGDLGLKGAKPQGHIGVDASAPQIGGSITGPSVEVQAPDIDVQGPGSKLNVPKMKVPKFSVSGAKGEETG IDVTLPTGEVTVPGVSGDVSLPEIATGGLEGKMKGTKVKTPEMIIQKPKISMQDVDLSLGSPKLKGDIKVSAPGVQGDVKGPQVALKGSR VDIETPNLEGTLTGPRLGSPSGKTGTCRISMSEVDLNVAAPKVKGGVDVTLPRVEGKVKVPEVDVRGPKVDVSAPDVEAHGPEWNLKMPK MKMPTFSTPGAKGEGPDVHMTLPKGDISISGPKVNVEAPDVNLEGLGGKLKGPDVKLPDMSVKTPKISMPDVDLHVKGTKVKGEYDVTVP KLEGELKGPKVDIDAPDVDVHGPDWHLKMPKMKMPKFSVPGFKAEGPEVDVNLPKADVDISGPKIDVTAPDVSIEEPEGKLKGPKFKMPE MNIKVPKISMPDVDLHLKGPNVKGEYDVTMPKVESEIKVPDVELKSAKMDIDVPDVEVQGPDWHLKMPKMKMPKFSMPGFKAEGPEVDVN LPKADVDISGPKVGVEVPDVNIEGPEGKLKGPKFKMPEMNIKAPKISMPDVDLHMKGPKVKGEYDMTVPKLEGDLKGPKVDVSAPDVEMQ GPDWNLKMPKIKMPKFSMPSLKGEGPEFDVNLSKANVDISAPKVDTNAPDLSLEGPEGKLKGPKFKMPEMHFRAPKMSLPDVDLDLKGPK MKGNVDISAPKIEGEMQVPDVDIRGPKVDIKAPDVEGQGLDWSLKIPKMKMPKFSMPSLKGEGPEVDVNLPKADVVVSGPKVDIEAPDVS LEGPEGKLKGPKFKMPEMHFKTPKISMPDVDLHLKGPKVKGDVDVSVPKVEGEMKVPDVEIKGPKMDIDAPDVEVQGPDWHLKMPKMKMP KFSMPGFKGEGREVDVNLPKADIDVSGPKVDVEVPDVSLEGPEGKLKGPKFKMPEMHFKAPKISMPDVDLNLKGPKLKGDVDVSLPEVEG EMKVPDVDIKGPKVDISAPDVDVHGPDWHLKMPKVKMPKFSMPGFKGEGPEVDVKLPKADVDVSGPKMDAEVPDVNIEGPDAKLKGPKFK MPEMSIKPQKISIPDVGLHLKGPKMKGDYDVTVPKVEGEIKAPDVDIKGPKVDINAPDVEVHGPDWHLKMPKVKMPKFSMPGFKGEGPEV DMNLPKADLGVSGPKVDIDVPDVNLEAPEGKLKGPKFKMPSMNIQTHKISMPDVGLNLKAPKLKTDVDVSLPKVEGDLKGPEIDVKAPKM DVNVGDIDIEGPEGKLKGPKFKMPEMHFKAPKISMPDVDLHLKGPKVKGDMDVSVPKVEGEMKVPDVDIKGPKVDIDAPDVEVHDPDWHL KMPKMKMPKFSMPGFKAEGPEVDVNLPKADIDVSGPSVDTDAPDLDIEGPEGKLKGSKFKMPKLNIKAPKVSMPDVDLNLKGPKLKGEID ASVPELEGDLRGPQVDVKGPFVEAEVPDVDLECPDAKLKGPKFKMPEMHFKAPKISMPDVDLHLKGPKVKGDADVSVPKLEGDLTGPSVG VEVPDVELECPDAKLKGPKFKMPDMHFKAPKISMPDVDLHLKGPKVKGDVDVSVPKLEGDLTGPSVGVEVPDVELECPDAKLKGPKFKMP EMHFKTPKISMPDVDLHLKGPKVKGDMDVSVPKVEGEMKVPDVDIKGPKMDIDAPDVDVHGPDWHLKMPKMKMPKFSMPGFKAEGPEVDV NLPKADVVVSGPKVDVEVPDVSLEGPEGKLKGPKLKMPEMHFKAPKISMPDVDLHLKGPKVKGDVDVSLPKLEGDLTGPSVDVEVPDVEL ECPDAKLKGPKFKMPEMHFKTPKISMPDVNLNLKGPKVKGDMDVSVPKVEGEMKVPDVDIRGPKVDIDAPDVDVHGPDWHLKMPKMKMPK FSMPGFKGEGPEVDVNLPKADVDVSGPKVDVEVPDVSLEGPEGKLKGPKFKMPEMHFKTPKISMPDVDFNLKGPKIKGDVDVSAPKLEGE LKGPELDVKGPKLDADMPEVAVEGPNGKWKTPKFKMPDMHFKAPKISMPDLDLHLKSPKAKGEVDVDVPKLEGDLKGPHVDVSGPDIDIE GPEGKLKGPKFKMPDMHFKAPNISMPDVDLNLKGPKIKGDVDVSVPEVEGKLEVPDMNIRGPKVDVNAPDVQAPDWHLKMPKMKMPKFSM PGFKAEGPEVDVNLPKADVDISGPKVDIEGPDVNIEGPEGKLKGPKLKMPEMNIKAPKISMPDFDLHLKGPKVKGDVDVSLPKVEGDLKG PEVDIKGPKVDINAPDVGVQGPDWHLKMPKVKMPKFSMPGFKGEGPDGDVKLPKADIDVSGPKVDIEGPDVNIEGPEGKLKGPKFKMPEM NIKAPKISMPDIDLNLKGPKVKGDVDVSLPKVEGDLKGPEVDIKGPKVDIDAPDVDVHGPDWHLKMPKIKMPKISMPGFKGEGPDVDVNL PKADIDVSGPKVDVECPDVNIEGPEGKWKSPKFKMPEMHFKTPKISMPDIDLNLTGPKIKGDVDVTGPKVEGDLKGPEVDLKGPKVDIDV PDVNVQGPDWHLKMPKMKMPKFSMPGFKAEGPEVDVNLPKADVDVSGPKVDVEGPDVNIEGPEGKLKGPKFKMPEMNIKAPKIPMPDFDL HLKGPKVKGDVDISLPKVEGDLKGPEVDIRGPQVDIDVPDVGVQGPDWHLKMPKVKMPKFSMPGFKGEGPDVDVNLPKADLDVSGPKVDI DVPDVNIEGPEGKLKGPKFKMPEMNIKAPKISMPDIDLNLKGPKVKGDMDVSLPKVEGDMKVPDVDIKGPKVDINAPDVDVQGPDWHLKM PKIKMPKISMPGFKGEGPEVDVNLPKADLDVSGPKVDVDVPDVNIEGPDAKLKGPKFKMPEMNIKAPKISMPDLDLNLKGPKMKGEVDVS LANVEGDLKGPALDIKGPKIDVDAPDIDIHGPDAKLKGPKLKMPDMHVNMPKISMPEIDLNLKGSKLKGDVDVSGPKLEGDIKAPSLDIK GPEVDVSGPKLNIEGKSKKSRFKLPKFNFSGSKVQTPEVDVKGKKPDIDITGPKVDINAPDVEVQGKVKGSKFKMPFLSISSPKVSMPDV ELNLKSPKVKGDLDIAGPNLEGDFKGPKVDIKAPEVNLNAPDVDVHGPDWNLKMPKMKMPKFSVSGLKAEGPDVAVDLPKGDINIEGPSM NIEGPDLNVEGPEGGLKGPKFKMPDMNIKAPKISMPDIDLNLKGPKVKGDVDISLPKLEGDLKGPEVDIKGPKVDINAPDVDVHGPDWHL KMPKVKMPKFSMPGFKGEGPEVDVTLPKADIDISGPNVDVDVPDVNIEGPDAKLKGPKFKMPEMNIKAPKISMPDFDLNLKGPKMKGDVV VSLPKVEGDLKGPEVDIKGPKVDIDTPDINIEGSEGKFKGPKFKIPEMHLKAPKISMPDIDLNLKGPKVKGDVDVSLPKMEGDLKGPEVD IKGPKVDINAPDVDVQGPDWHLKMPKVKMPKFSMPGFKGEGPDVDVNLPKADLDVSGPKVDIDVPDVNIEGPEGKLKGPKFKMPEMNIKA PKISMPDIDLNLKGPKVKGDMDVSLPKVEGDMQVPDLDIKGPKVDINAPDVDVRGPDWHLKMPKIKMPKISMPGFKGEGPEVDVNLPKAD LDVSGPKVDVDVPDVNIEGPDAKLKGPKFKMPEMNIKAPKISMPDFDLHLKGPKVKGDVDVSLPKMEGDLKAPEVDIKGPKVDIDAPDVD VHGPDWHLKMPKVKMPKFSMPGFKGEGPEVDVNLPKADIDVSGPKVDIDTPDIDIHGPEGKLKGPKFKMPDLHLKAPKISMPEVDLNLKG PKMKGDVDVSLPKVEGDLKGPEVDIKGPKVDIDVPDVDVQGPDWHLKMPKVKMPKFSMPGFKGEGPDVDVNLPKADLDVSGPKVDIDVPD VNIEGPDAKLKGPKFKMPEMNIKAPKISMPDFDLHLKGPKVKGDVDVSLPKVEGDLKGPEVDIKGPKVDIDAPDVDVHGPDWHLKMPKVK MPKFSMPGFKGEGPDVDVTLPKADIEISGPKVDIDAPDVSIEGPDAKLKGPKFKMPEMNIKAPKISMPDIDFNLKGPKVKGDVDVSLPKV EGDLKGPEIDIKGPSLDIDTPDVNIEGPEGKLKGPKFKMPEMNIKAPKISMPDFDLHLKGPKVKGDVDVSLPKVESDLKGPEVDIEGPEG KLKGPKFKMPDVHFKSPQISMSDIDLNLKGPKIKGDMDISVPKLEGDLKGPKVDVKGPKVGIDTPDIDIHGPEGKLKGPKFKMPDLHLKA PKISMPEVDLNLKGPKVKGDMDISLPKVEGDLKGPEVDIRDPKVDIDVPDVDVQGPDWHLKMPKVKMPKFSMPGFKGEGPDVDVNLPKAD IDVSGPKVDVDVPDVNIEGPDAKLKGPKFKMPEMSIKAPKISMPDIDLNLKGPKVKGDVDVTLPKVEGDLKGPEADIKGPKVDINTPDVD VHGPDWHLKMPKVKMPKFSMPGFKGEGPDVDVSLPKADIDVSGPKVDVDIPDVNIEGPDAKLKGPKFKMPEINIKAPKISIPDVDLDLKG PKVKGDFDVSVPKVEGTLKGPEVDLKGPRLDFEGPDAKLSGPSLKMPSLEISAPKVTAPDVDLHLKAPKIGFSGPKLEGGEVDLKGPKVE APSLDVHMDSPDINIEGPDVKIPKFKKPKFGFGAKSPKADIKSPSLDVTVPEAELNLETPEISVGGKGKKSKFKMPKIHMSGPKIKAKKQ GFDLNVPGGEIDASLKAPDVDVNIAGPDAALKVDVKSPKTKKTMFGKMYFPDVEFDIKSPKFKAEAPLPSPKLEGELQAPDLELSLPAIH VEGLDIKAKAPKVKMPDVDISVPKIEGDLKGPKVQANLGAPDINIEGLDAKVKTPSFGISAPQVSIPDVNVNLKGPKIKGDVPSVGLEGP DVDLQGPEAKIKFPKFSMPKIGIPGVKMEGGGAEVHAQLPSLEGDLRGPDVKLEGPDVSLKGPGVDLPSVNLSMPKVSGPDLDLNLKGPS LKGDLDASVPSMKVHAPGLNLSGVGGKMQVGGDGVKVPGIDATTKLNVGAPDVTLRGPSLQGDLAVSGDIKCPKVSVGAPDLSLEASEGS IKLPKMKLPQFGISTPGSDLHVNAKGPQVSGELKGPGVDVNLKGPRISAPNVDFNLEGPKVKGSLGATGEIKGPTVGGGLPGIGVQGLEG NLQMPGIKSSGCDVNLPGVNVKLPTGQISGPEIKGGLKGSEVGFHGAAPDISVKGPAFNMASPESDFGINLKGPKIKGGADVSGGVSAPD ISLGEGHLSVKGSGGEWKGPQVSSALNLDTSKFAGGLHFSGPKVEGGVKGGQIGLQAPGLSVSGPQGHLESGSGKVTFPKMKIPKFTFSG RELVGREMGVDVHFPKAEASIQAGAGDGEWEESEVKLKKSKIKMPKFNFSKPKGKGGVTGSPEASISGSKGDLKSSKASLGSLEGEAEAE ASSPKGKFSLFKSKKPRHRSNSFSDEREFSGPSTPTGTLEFEGGEVSLEGGKVKGKHGKLKFGTFGGLGSKSKGHYEVTGSDDETGKLQG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:62283375/chr2:32820101) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AHNAK | BIRC6 |
| FUNCTION: Anti-apoptotic protein which can regulate cell death by controlling caspases and by acting as an E3 ubiquitin-protein ligase. Has an unusual ubiquitin conjugation system in that it could combine in a single polypeptide, ubiquitin conjugating (E2) with ubiquitin ligase (E3) activity, forming a chimeric E2/E3 ubiquitin ligase. Its tragets include CASP9 and DIABLO/SMAC. Acts as an inhibitor of CASP3, CASP7 and CASP9. Important regulator for the final stages of cytokinesis. Crucial for normal vesicle targeting to the site of abscission, but also for the integrity of the midbody and the midbody ring, and its striking ubiquitin modification. {ECO:0000269|PubMed:14765125, ECO:0000269|PubMed:15200957, ECO:0000269|PubMed:18329369}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 5458_5654 | 6170.666666666667 | 5891.0 | Compositional bias | Note=Gly-rich |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 9_90 | 6170.666666666667 | 5891.0 | Domain | PDZ |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 4971_4979 | 6170.666666666667 | 5891.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 5019_5027 | 6170.666666666667 | 5891.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 5034_5039 | 6170.666666666667 | 5891.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 5706_5716 | 6170.666666666667 | 5891.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000378024 | - | 5 | 5 | 5772_5779 | 6170.666666666667 | 5891.0 | Motif | Nuclear localization signal |
| Tgene | BIRC6 | chr11:62283375 | chr2:32820101 | ENST00000421745 | 66 | 74 | 4573_4740 | 4500.333333333333 | 4858.0 | Domain | UBC core |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 5458_5654 | 0 | 150.0 | Compositional bias | Note=Gly-rich |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 9_90 | 0 | 150.0 | Domain | PDZ |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 4971_4979 | 0 | 150.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 5019_5027 | 0 | 150.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 5034_5039 | 0 | 150.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 5706_5716 | 0 | 150.0 | Motif | Nuclear localization signal |
| Hgene | AHNAK | chr11:62283375 | chr2:32820101 | ENST00000257247 | - | 1 | 6 | 5772_5779 | 0 | 150.0 | Motif | Nuclear localization signal |
| Tgene | BIRC6 | chr11:62283375 | chr2:32820101 | ENST00000421745 | 66 | 74 | 1660_1668 | 4500.333333333333 | 4858.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BIRC6 | chr11:62283375 | chr2:32820101 | ENST00000421745 | 66 | 74 | 30_36 | 4500.333333333333 | 4858.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | BIRC6 | chr11:62283375 | chr2:32820101 | ENST00000421745 | 66 | 74 | 284_358 | 4500.333333333333 | 4858.0 | Repeat | Note=BIR |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| AHNAK | |
| BIRC6 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to AHNAK-BIRC6 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to AHNAK-BIRC6 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

