| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HIPK2-BRAF |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HIPK2-BRAF | FusionPDB ID: 36401 | FusionGDB2.0 ID: 36401 | Hgene | Tgene | Gene symbol | HIPK2 | BRAF | Gene ID | 28996 | 673 |
| Gene name | homeodomain interacting protein kinase 2 | B-Raf proto-oncogene, serine/threonine kinase | |
| Synonyms | PRO0593 | B-RAF1|B-raf|BRAF1|NS7|RAFB1 | |
| Cytomap | 7q34 | 7q34 | |
| Type of gene | protein-coding | protein-coding | |
| Description | homeodomain-interacting protein kinase 2hHIPk2 | serine/threonine-protein kinase B-raf94 kDa B-raf proteinB-Raf proto-oncogene serine/threonine-protein kinase (p94)B-Raf serine/threonine-proteinmurine sarcoma viral (v-raf) oncogene homolog B1proto-oncogene B-Rafv-raf murine sarcoma viral oncogene | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q9H2X6 | P15056 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000342645, ENST00000406875, ENST00000428878, | ENST00000288602, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 17 X 13 X 12=2652 | 48 X 58 X 16=44544 |
| # samples | 25 | 69 | |
| ** MAII score | log2(25/2652*10)=-3.4070807754505 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(69/44544*10)=-6.0124909441832 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HIPK2 [Title/Abstract] AND BRAF [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HIPK2(139297947)-BRAF(140449218), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HIPK2-BRAF seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HIPK2-BRAF seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HIPK2-BRAF seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HIPK2-BRAF seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HIPK2 | GO:0006468 | protein phosphorylation | 19448668 |
| Hgene | HIPK2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 14647468 |
| Hgene | HIPK2 | GO:0045766 | positive regulation of angiogenesis | 19046997 |
| Hgene | HIPK2 | GO:0060395 | SMAD protein signal transduction | 12874272 |
| Tgene | BRAF | GO:0000186 | activation of MAPKK activity | 29433126 |
| Tgene | BRAF | GO:0006468 | protein phosphorylation | 17563371 |
| Tgene | BRAF | GO:0010828 | positive regulation of glucose transmembrane transport | 23010278 |
| Tgene | BRAF | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19667065 |
| Tgene | BRAF | GO:0043066 | negative regulation of apoptotic process | 19667065 |
| Tgene | BRAF | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 22065586 |
| Tgene | BRAF | GO:0071277 | cellular response to calcium ion | 18567582 |
| Tgene | BRAF | GO:0090150 | establishment of protein localization to membrane | 23010278 |
 Fusion gene breakpoints across HIPK2 (5'-gene) Fusion gene breakpoints across HIPK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
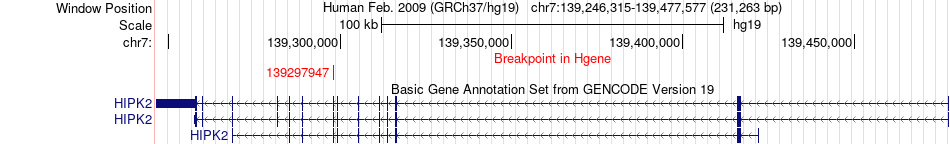 |
 Fusion gene breakpoints across BRAF (3'-gene) Fusion gene breakpoints across BRAF (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
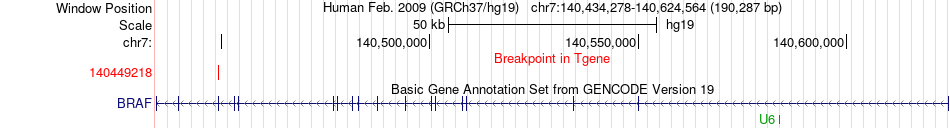 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-VQ-A92D | HIPK2 | chr7 | 139297947 | - | BRAF | chr7 | 140449218 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000406875 | HIPK2 | chr7 | 139297947 | - | ENST00000288602 | BRAF | chr7 | 140449218 | - | 2766 | 2207 | 95 | 2647 | 850 |
| ENST00000428878 | HIPK2 | chr7 | 139297947 | - | ENST00000288602 | BRAF | chr7 | 140449218 | - | 2745 | 2186 | 155 | 2626 | 823 |
| ENST00000342645 | HIPK2 | chr7 | 139297947 | - | ENST00000288602 | BRAF | chr7 | 140449218 | - | 2671 | 2112 | 21 | 2552 | 843 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000406875 | ENST00000288602 | HIPK2 | chr7 | 139297947 | - | BRAF | chr7 | 140449218 | - | 0.004015429 | 0.99598455 |
| ENST00000428878 | ENST00000288602 | HIPK2 | chr7 | 139297947 | - | BRAF | chr7 | 140449218 | - | 0.005234031 | 0.994766 |
| ENST00000342645 | ENST00000288602 | HIPK2 | chr7 | 139297947 | - | BRAF | chr7 | 140449218 | - | 0.003625919 | 0.9963741 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >36401_36401_1_HIPK2-BRAF_HIPK2_chr7_139297947_ENST00000342645_BRAF_chr7_140449218_ENST00000288602_length(amino acids)=843AA_BP=691 MASHVQVFSPHTLQSSAFCSVKKLKIEPSSNWDMTGYGSHSKVYSQSKNIPLSQPATTTVSTSLPVPNPSLPYEQTIVFPGSTGHIVVTS ASSTSVTGQVLGGPHNLMRRSTVSLLDTYQKCGLKRKSEEIENTSSVQIIEEHPPMIQNNASGATVATATTSTATSKNSGSNSEGDYQLV QHEVLCSMTNTYEVLEFLGRGTFGQVVKCWKRGTNEIVAIKILKNHPSYARQGQIEVSILARLSTESADDYNFVRAYECFQHKNHTCLVF EMLEQNLYDFLKQNKFSPLPLKYIRPVLQQVATALMKLKSLGLIHADLKPENIMLVDPSRQPYRVKVIDFGSASHVSKAVCSTYLQSRYY RAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLSAGTKTTRFFNRDTDSPYPLWRLKTPDDHEAE TGIKSKEARKYIFNCLDDMAQVNMTTDLEGSDMLVEKADRREFIDLLKKMLTIDADKRITPIETLNHPFVTMTHLLDFPHSTHVKSCFQN MEICKRRVNMYDTVNQSKTPFITHVAPSTSTNLTMTFNNQLTTVHNQAPSSTSATISLANPEVSILNYPSTLYQPSAASMAAVAQRSMPL QTGTAQICARPDPFQQALIVCPPGFQGLQASPSKHAGYSVRMENAVPIVTQAPGAQPLQIQPGLLAQAPEVIRMQDKNPYSFQSDVYAFG IVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHRSASEPSL -------------------------------------------------------------- >36401_36401_2_HIPK2-BRAF_HIPK2_chr7_139297947_ENST00000406875_BRAF_chr7_140449218_ENST00000288602_length(amino acids)=850AA_BP=698 MAPVYEGMASHVQVFSPHTLQSSAFCSVKKLKIEPSSNWDMTGYGSHSKVYSQSKNIPLSQPATTTVSTSLPVPNPSLPYEQTIVFPGST GHIVVTSASSTSVTGQVLGGPHNLMRRSTVSLLDTYQKCGLKRKSEEIENTSSVQIIEEHPPMIQNNASGATVATATTSTATSKNSGSNS EGDYQLVQHEVLCSMTNTYEVLEFLGRGTFGQVVKCWKRGTNEIVAIKILKNHPSYARQGQIEVSILARLSTESADDYNFVRAYECFQHK NHTCLVFEMLEQNLYDFLKQNKFSPLPLKYIRPVLQQVATALMKLKSLGLIHADLKPENIMLVDPSRQPYRVKVIDFGSASHVSKAVCST YLQSRYYRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLSAGTKTTRFFNRDTDSPYPLWRLKT PDDHEAETGIKSKEARKYIFNCLDDMAQVNMTTDLEGSDMLVEKADRREFIDLLKKMLTIDADKRITPIETLNHPFVTMTHLLDFPHSTH VKSCFQNMEICKRRVNMYDTVNQSKTPFITHVAPSTSTNLTMTFNNQLTTVHNQAPSSTSATISLANPEVSILNYPSTLYQPSAASMAAV AQRSMPLQTGTAQICARPDPFQQALIVCPPGFQGLQASPSKHAGYSVRMENAVPIVTQAPGAQPLQIQPGLLAQAPEVIRMQDKNPYSFQ SDVYAFGIVLYELMTGQLPYSNINNRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHR -------------------------------------------------------------- >36401_36401_3_HIPK2-BRAF_HIPK2_chr7_139297947_ENST00000428878_BRAF_chr7_140449218_ENST00000288602_length(amino acids)=823AA_BP=671 MAPVYEGMASHVQVFSPHTLQSSAFCSVKKLKIEPSSNWDMTGYGSHSKVYSQSKNIPLSQPATTTVSTSLPVPNPSLPYEQTIVFPGST GHIVVTSASSTSVTGQVLGGPHNLMRRSTVSLLDTYQKCGLKRKSEEIENTSSVQIIEEHPPMIQNNASGATVATATTSTATSKNSGSNS EGDYQLVQHEVLCSMTNTYEVLEFLGRGTFGQVVKCWKRGTNEIVAIKILKNHPSYARQGQIEVSILARLSTESADDYNFVRAYECFQHK NHTCLVFEMLEQNLYDFLKQNKFSPLPLKYIRPVLQQVATALMKLKSLGLIHADLKPENIMLVDPSRQPYRVKVIDFGSASHVSKAVCST YLQSRYYRAPEIILGLPFCEAIDMWSLGCVIAELFLGWPLYPGASEYDQIRYISQTQGLPAEYLLSAGTKTTRFFNRDTDSPYPLWRLKT PDDHEAETGIKSKEARKYIFNCLDDMAQVNMTTDLEGSDMLVEKADRREFIDLLKKMLTIDADKRITPIETLNHPFVTMTHLLDFPHSTH VKSCFQNMEICKRRVNMYDTVNQSKTPFITHVAPSTSTNLTMTFNNQLTTVHNQPSAASMAAVAQRSMPLQTGTAQICARPDPFQQALIV CPPGFQGLQASPSKHAGYSVRMENAVPIVTQAPGAQPLQIQPGLLAQAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNINNRD QIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARSLPKIHRSASEPSLNRAGFQTEDFSLYACASPKT -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:139297947/chr7:140449218) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HIPK2 | BRAF |
| FUNCTION: Serine/threonine-protein kinase involved in transcription regulation, p53/TP53-mediated cellular apoptosis and regulation of the cell cycle. Acts as a corepressor of several transcription factors, including SMAD1 and POU4F1/Brn3a and probably NK homeodomain transcription factors. Phosphorylates PDX1, ATF1, PML, p53/TP53, CREB1, CTBP1, CBX4, RUNX1, EP300, CTNNB1, HMGA1 and ZBTB4. Inhibits cell growth and promotes apoptosis through the activation of p53/TP53 both at the transcription level and at the protein level (by phosphorylation and indirect acetylation). The phosphorylation of p53/TP53 may be mediated by a p53/TP53-HIPK2-AXIN1 complex. Involved in the response to hypoxia by acting as a transcriptional co-suppressor of HIF1A. Mediates transcriptional activation of TP73. In response to TGFB, cooperates with DAXX to activate JNK. Negative regulator through phosphorylation and subsequent proteasomal degradation of CTNNB1 and the antiapoptotic factor CTBP1. In the Wnt/beta-catenin signaling pathway acts as an intermediate kinase between MAP3K7/TAK1 and NLK to promote the proteasomal degradation of MYB. Phosphorylates CBX4 upon DNA damage and promotes its E3 SUMO-protein ligase activity. Activates CREB1 and ATF1 transcription factors by phosphorylation in response to genotoxic stress. In response to DNA damage, stabilizes PML by phosphorylation. PML, HIPK2 and FBXO3 may act synergically to activate p53/TP53-dependent transactivation. Promotes angiogenesis, and is involved in erythroid differentiation, especially during fetal liver erythropoiesis. Phosphorylation of RUNX1 and EP300 stimulates EP300 transcription regulation activity. Triggers ZBTB4 protein degradation in response to DNA damage. Modulates HMGA1 DNA-binding affinity. In response to high glucose, triggers phosphorylation-mediated subnuclear localization shifting of PDX1. Involved in the regulation of eye size, lens formation and retinal lamination during late embryogenesis. {ECO:0000269|PubMed:11740489, ECO:0000269|PubMed:11925430, ECO:0000269|PubMed:12851404, ECO:0000269|PubMed:12874272, ECO:0000269|PubMed:14678985, ECO:0000269|PubMed:17018294, ECO:0000269|PubMed:17960875, ECO:0000269|PubMed:18695000, ECO:0000269|PubMed:18809579, ECO:0000269|PubMed:19015637, ECO:0000269|PubMed:19046997, ECO:0000269|PubMed:19448668, ECO:0000269|PubMed:20307497, ECO:0000269|PubMed:20573984, ECO:0000269|PubMed:20637728, ECO:0000269|PubMed:20980392, ECO:0000269|PubMed:21192925, ECO:0000269|PubMed:22825850}. | FUNCTION: Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway (PubMed:21441910, PubMed:29433126). May play a role in the postsynaptic responses of hippocampal neurons (PubMed:1508179). {ECO:0000269|PubMed:1508179, ECO:0000269|PubMed:21441910, ECO:0000269|PubMed:29433126, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 199_527 | 704.0 | 1199.0 | Domain | Protein kinase |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 199_527 | 677.0 | 1172.0 | Domain | Protein kinase |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 205_213 | 704.0 | 1199.0 | Nucleotide binding | ATP |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 205_213 | 677.0 | 1172.0 | Nucleotide binding | ATP |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 97_230 | 704.0 | 1199.0 | Region | Transcriptional corepression |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 97_230 | 677.0 | 1172.0 | Region | Transcriptional corepression |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 1088_1094 | 704.0 | 1199.0 | Compositional bias | Note=Poly-Ala |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 1088_1094 | 677.0 | 1172.0 | Compositional bias | Note=Poly-Ala |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 802_805 | 704.0 | 1199.0 | Motif | Note=Nuclear localization signal 1 (NLS1) |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 832_835 | 704.0 | 1199.0 | Motif | Note=Nuclear localization signal 2 (NLS2) |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 802_805 | 677.0 | 1172.0 | Motif | Note=Nuclear localization signal 1 (NLS1) |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 832_835 | 677.0 | 1172.0 | Motif | Note=Nuclear localization signal 2 (NLS2) |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 873_980 | 704.0 | 1199.0 | Region | Required for localization to nuclear speckles |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 984_1198 | 704.0 | 1199.0 | Region | Note=Autoinhibitory domain (AID) |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 873_980 | 677.0 | 1172.0 | Region | Required for localization to nuclear speckles |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 984_1198 | 677.0 | 1172.0 | Region | Note=Autoinhibitory domain (AID) |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 122_129 | 620.0 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 428_432 | 620.0 | 767.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 6_11 | 620.0 | 767.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 155_227 | 620.0 | 767.0 | Domain | RBD | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 457_717 | 620.0 | 767.0 | Domain | Protein kinase | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 463_471 | 620.0 | 767.0 | Nucleotide binding | ATP | |
| Tgene | BRAF | chr7:139297947 | chr7:140449218 | ENST00000288602 | 14 | 18 | 234_280 | 620.0 | 767.0 | Zinc finger | Phorbol-ester/DAG-type |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| BRAF | YWHAB, YWHAG, YWHAQ, YWHAZ, SFN, HRAS, AKT1, MAPK3, RAP1GAP, RAF1, MRAS, RAP1A, PAK2, TERF1, CCDC88A, NEDD4L, Nedd4, MAP2K1, RNF149, KSR1, BRAP, PRKCE, RPS6KB2, HSP90AA1, BRAF, YWHAE, HSPA5, MAP2K2, HSPA1A, HSPA8, YWHAH, HSPA9, ARAF, CDC37, HSP90AB1, PHKB, LIMK1, IQGAP1, MAPK1, BAD, LIPF, MUS81, Vps4b, FBXW7, FGFR2, BRCA2, VHL, FNTA, HDAC2, PIK3CA, EGFR, PTEN, FNIP1, FNIP2, RAB3GAP1, KRAS, KIAA0141, FKBPL, ARMCX3, KCNC4, RPTOR, CYLD, NRAS, HSPA4, DNAJB6, PDCD11, PIP5K1A, DNAJC15, VANGL1, DNAJC11, FKBP5, HSPA4L, HSP90B1, GNAI2, DNAJC13, GNAS, PPP2CB, HSD11B2, DNAJB11, PLD2, RAP1B, WDR6, CPNE3, MYOF, COPA, UBLCP1, PPP1CA, RAD50, PIP4K2C, PHB, VIM, PGAM1, DNAJA1, MAP2K7, SPRY2, ALDOA, AP2B1, ATP5A1, SSB, IGF1R, JUP, PPP6C, PARP1, HSPB1, NME2, PRDX2, CCT7, RAB1A, FARSA, FASN, EPRS, TRAF2, REST, KIAA1429, NANOG, ITCH, SMURF2, WWP1, WWP2, PPP2CA, PPP2R2A, AURKA, LATS2, MAP2K3, MAP2K6, RASSF1, STK11, TERT, PEBP1, CRBN, FAR1, PSMC4, UBA52, UBB, UBC, RPS27A, HSPA6, HSP90AB3P, DSP, ATAD3A, ATAD3B, P4HB, SDF2L1, SLC25A22, SPTBN4, TMEM33, CTSB, NCL, HPX, TXNDC12, SLC25A11, NDUFA4, CTSV, FBP1, HSD17B3, ZNF189, ZNF510, KIF14, SRC, TRAP1, JTB, S100P, USP28, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HIPK2 | |
| BRAF | 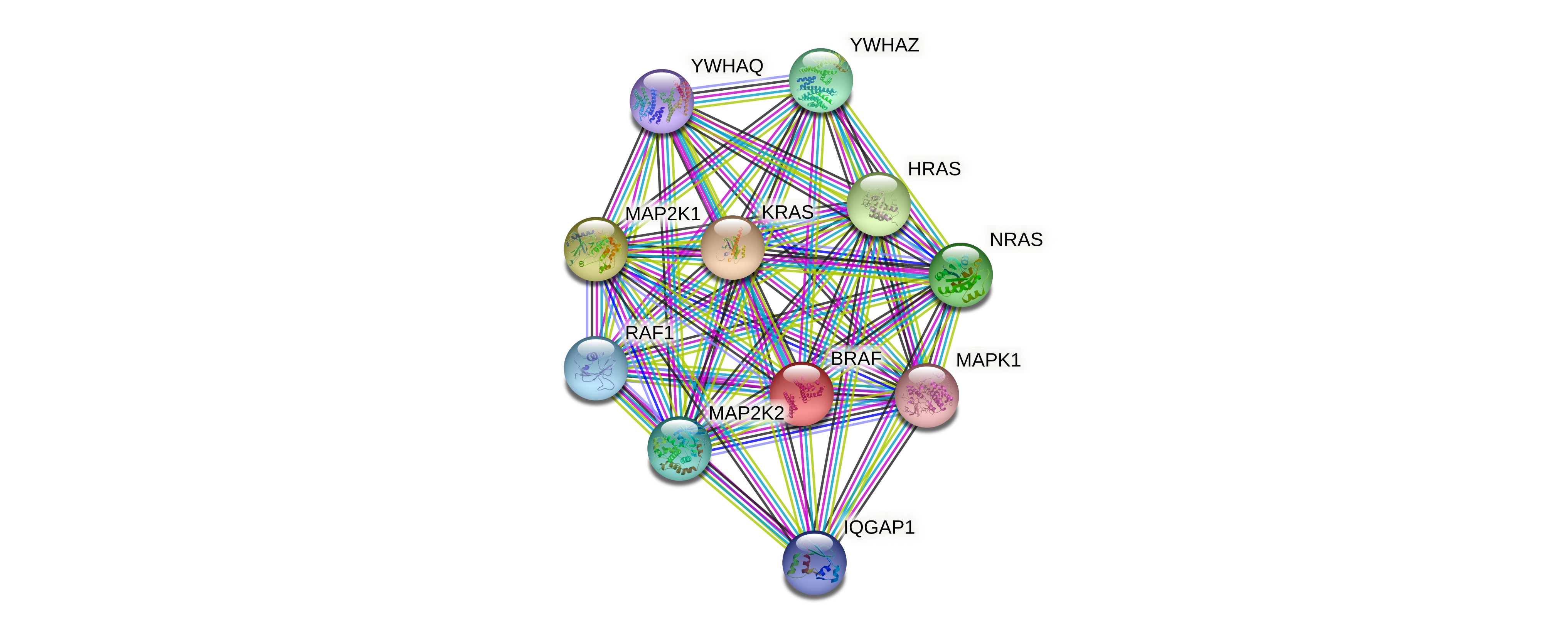 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 935_1049 | 704.0 | 1199.0 | AXIN1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 935_1049 | 677.0 | 1172.0 | AXIN1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 774_876 | 704.0 | 1199.0 | CTBP1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 774_876 | 677.0 | 1172.0 | CTBP1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 787_897 | 704.0 | 1199.0 | HMGA1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 787_897 | 677.0 | 1172.0 | HMGA1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 752_897 | 704.0 | 1199.0 | POU4F1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 752_897 | 677.0 | 1172.0 | POU4F1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 539_844 | 704.0 | 1199.0 | SKI and SMAD1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 539_844 | 677.0 | 1172.0 | SKI and SMAD1 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 846_941 | 704.0 | 1199.0 | TP53 and TP73 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 846_941 | 677.0 | 1172.0 | TP53 and TP73 |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000406875 | - | 9 | 15 | 873_907 | 704.0 | 1199.0 | UBE2I |
| Hgene | HIPK2 | chr7:139297947 | chr7:140449218 | ENST00000428878 | - | 9 | 15 | 873_907 | 677.0 | 1172.0 | UBE2I |
Top |
Related Drugs to HIPK2-BRAF |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HIPK2-BRAF |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | BRAF | C0025202 | melanoma | 24 | CGI;CTD_human;UNIPROT |
| Tgene | BRAF | C1275081 | Cardio-facio-cutaneous syndrome | 14 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | BRAF | C0009402 | Colorectal Carcinoma | 8 | CTD_human;UNIPROT |
| Tgene | BRAF | C0028326 | Noonan Syndrome | 8 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | BRAF | C0238463 | Papillary thyroid carcinoma | 8 | CTD_human;ORPHANET |
| Tgene | BRAF | C0040136 | Thyroid Neoplasm | 6 | CGI;CTD_human |
| Tgene | BRAF | C0151468 | Thyroid Gland Follicular Adenoma | 6 | CTD_human |
| Tgene | BRAF | C0175704 | LEOPARD Syndrome | 6 | CLINGEN;GENOMICS_ENGLAND |
| Tgene | BRAF | C0549473 | Thyroid carcinoma | 6 | CGI;CTD_human |
| Tgene | BRAF | C3150970 | NOONAN SYNDROME 7 | 5 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | BRAF | C0009404 | Colorectal Neoplasms | 4 | CTD_human |
| Tgene | BRAF | C3150971 | LEOPARD SYNDROME 3 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | BRAF | C1519086 | Pilomyxoid astrocytoma | 3 | ORPHANET |
| Tgene | BRAF | C0004565 | Melanoma, B16 | 2 | CTD_human |
| Tgene | BRAF | C0009075 | Melanoma, Cloudman S91 | 2 | CTD_human |
| Tgene | BRAF | C0018598 | Melanoma, Harding-Passey | 2 | CTD_human |
| Tgene | BRAF | C0023443 | Hairy Cell Leukemia | 2 | CGI;ORPHANET |
| Tgene | BRAF | C0025205 | Melanoma, Experimental | 2 | CTD_human |
| Tgene | BRAF | C0033578 | Prostatic Neoplasms | 2 | CTD_human |
| Tgene | BRAF | C0152013 | Adenocarcinoma of lung (disorder) | 2 | CGI;CTD_human |
| Tgene | BRAF | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human |
| Tgene | BRAF | C0587248 | Costello syndrome (disorder) | 2 | CLINGEN;CTD_human |
| Tgene | BRAF | C3501843 | Nonmedullary Thyroid Carcinoma | 2 | CTD_human |
| Tgene | BRAF | C3501844 | Familial Nonmedullary Thyroid Cancer | 2 | CTD_human |
| Tgene | BRAF | C0002448 | Ameloblastoma | 1 | CTD_human |
| Tgene | BRAF | C0004114 | Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0010276 | Craniopharyngioma | 1 | CTD_human;ORPHANET |
| Tgene | BRAF | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Tgene | BRAF | C0017638 | Glioma | 1 | CGI;CTD_human |
| Tgene | BRAF | C0019621 | Histiocytosis, Langerhans-Cell | 1 | CGI;ORPHANET |
| Tgene | BRAF | C0022665 | Kidney Neoplasm | 1 | CTD_human |
| Tgene | BRAF | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | BRAF | C0024232 | Lymphatic Metastasis | 1 | CTD_human |
| Tgene | BRAF | C0024694 | Mandibular Neoplasms | 1 | CTD_human |
| Tgene | BRAF | C0027659 | Neoplasms, Experimental | 1 | CTD_human |
| Tgene | BRAF | C0027962 | Melanocytic nevus | 1 | GENOMICS_ENGLAND |
| Tgene | BRAF | C0036920 | Sezary Syndrome | 1 | CTD_human |
| Tgene | BRAF | C0041409 | Turner Syndrome, Male | 1 | CTD_human |
| Tgene | BRAF | C0079773 | Lymphoma, T-Cell, Cutaneous | 1 | CTD_human |
| Tgene | BRAF | C0205768 | Subependymal Giant Cell Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0206686 | Adrenocortical carcinoma | 1 | CTD_human |
| Tgene | BRAF | C0206754 | Neuroendocrine Tumors | 1 | CTD_human |
| Tgene | BRAF | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | BRAF | C0278875 | Adult Craniopharyngioma | 1 | CTD_human |
| Tgene | BRAF | C0280783 | Juvenile Pilocytic Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0280785 | Diffuse Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0334579 | Anaplastic astrocytoma | 1 | CGI;CTD_human |
| Tgene | BRAF | C0334580 | Protoplasmic astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0334581 | Gemistocytic astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0334582 | Fibrillary Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0334583 | Pilocytic Astrocytoma | 1 | CGI;CTD_human |
| Tgene | BRAF | C0338070 | Childhood Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | BRAF | C0376407 | Granulomatous Slack Skin | 1 | CTD_human |
| Tgene | BRAF | C0406803 | Syringocystadenoma Papilliferum | 1 | GENOMICS_ENGLAND |
| Tgene | BRAF | C0431128 | Papillary craniopharyngioma | 1 | CTD_human |
| Tgene | BRAF | C0431129 | Adamantinous Craniopharyngioma | 1 | CTD_human |
| Tgene | BRAF | C0547065 | Mixed oligoastrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | BRAF | C0596263 | Carcinogenesis | 1 | CTD_human |
| Tgene | BRAF | C0684249 | Carcinoma of lung | 1 | CGI;UNIPROT |
| Tgene | BRAF | C0740457 | Malignant neoplasm of kidney | 1 | CTD_human |
| Tgene | BRAF | C0750935 | Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0750936 | Intracranial Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C0751061 | Craniopharyngioma, Child | 1 | CTD_human |
| Tgene | BRAF | C0920269 | Microsatellite Instability | 1 | CTD_human |
| Tgene | BRAF | C1527404 | Female Pseudo-Turner Syndrome | 1 | CTD_human |
| Tgene | BRAF | C1704230 | Grade I Astrocytoma | 1 | CTD_human |
| Tgene | BRAF | C1721098 | Replication Error Phenotype | 1 | CTD_human |
| Tgene | BRAF | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | BRAF | C4551484 | Leopard Syndrome 1 | 1 | GENOMICS_ENGLAND |
| Tgene | BRAF | C4551602 | Noonan Syndrome 1 | 1 | CTD_human |
| Tgene | BRAF | C4721532 | Lymphoma, Non-Hodgkin, Familial | 1 | UNIPROT |
| Tgene | BRAF | C4733333 | familial non-medullary thyroid cancer | 1 | GENOMICS_ENGLAND |

