| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HIVEP3-LRP8 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HIVEP3-LRP8 | FusionPDB ID: 36558 | FusionGDB2.0 ID: 36558 | Hgene | Tgene | Gene symbol | HIVEP3 | LRP8 | Gene ID | 59269 | 7804 |
| Gene name | HIVEP zinc finger 3 | LDL receptor related protein 8 | |
| Synonyms | KBP-1|KBP1|KRC|SHN3|Schnurri-3|ZAS3|ZNF40C | APOER2|HSZ75190|LRP-8|MCI1 | |
| Cytomap | 1p34.2 | 1p32.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcription factor HIVEP3ZAS family, member 3human immunodeficiency virus type I enhancer binding protein 3kappa-B and V(D)J recombination signal sequences-binding proteinkappa-binding protein 1zinc finger protein ZAS3 | low-density lipoprotein receptor-related protein 8ApoE receptor 2low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q5T1R4 | Q14114 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000247584, ENST00000372583, ENST00000372584, ENST00000429157, ENST00000460604, | ENST00000347547, ENST00000354412, ENST00000465675, ENST00000460214, ENST00000306052, ENST00000371454, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 21 X 18 X 10=3780 | 4 X 3 X 4=48 |
| # samples | 25 | 4 | |
| ** MAII score | log2(25/3780*10)=-3.91838623444635 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HIVEP3 [Title/Abstract] AND LRP8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HIVEP3(42041214)-LRP8(53792664), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. HIVEP3-LRP8 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HIVEP3 | GO:0045893 | positive regulation of transcription, DNA-templated | 15790681 |
| Tgene | LRP8 | GO:0006897 | endocytosis | 8626535 |
 Fusion gene breakpoints across HIVEP3 (5'-gene) Fusion gene breakpoints across HIVEP3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
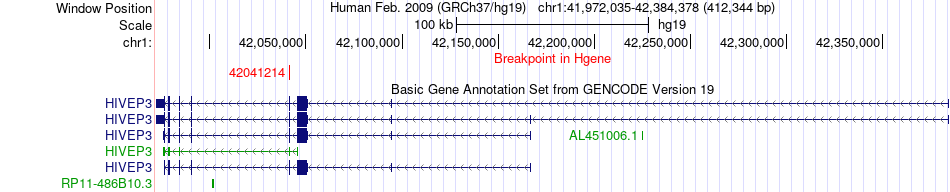 |
 Fusion gene breakpoints across LRP8 (3'-gene) Fusion gene breakpoints across LRP8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
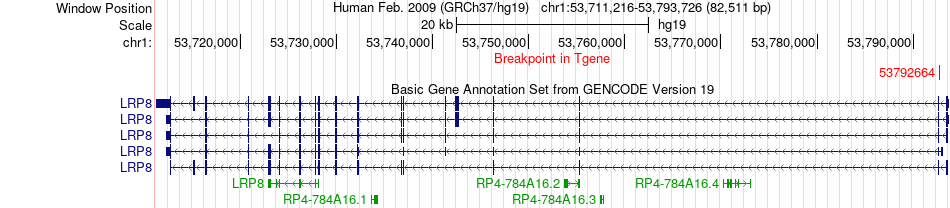 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-IG-A51D | HIVEP3 | chr1 | 42041214 | - | LRP8 | chr1 | 53792664 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000372583 | HIVEP3 | chr1 | 42041214 | - | ENST00000306052 | LRP8 | chr1 | 53792664 | - | 10334 | 6093 | 886 | 6180 | 1764 |
| ENST00000372583 | HIVEP3 | chr1 | 42041214 | - | ENST00000371454 | LRP8 | chr1 | 53792664 | - | 9149 | 6093 | 886 | 6180 | 1764 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000372583 | ENST00000306052 | HIVEP3 | chr1 | 42041214 | - | LRP8 | chr1 | 53792664 | - | 0.005030297 | 0.99496967 |
| ENST00000372583 | ENST00000371454 | HIVEP3 | chr1 | 42041214 | - | LRP8 | chr1 | 53792664 | - | 0.007761939 | 0.9922381 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >36558_36558_1_HIVEP3-LRP8_HIVEP3_chr1_42041214_ENST00000372583_LRP8_chr1_53792664_ENST00000306052_length(amino acids)=1764AA_BP=1736 MDPEQSVKGTKKAEGSPRKRLTKGEAIQTSVSSSVPYPGSGTAATQESPAQELLAPQPFPGPSSVLREGSQEKTGQQQKPPKRPPIEASV HISQLPQHPLTPAFMSPGKPEHLLEGSTWQLVDPMRPGPSGSFVAPGLHPQSQLLPSHASIIPPEDLPGVPKVFVPRPSQVSLKPTEEAH KKERKPQKPGKYICQYCSRPCAKPSVLQKHIRSHTGERPYPCGPCGFSFKTKSNLYKHRKSHAHRIKAGLASGMGGEMYPHGLEMERIPG EEFEEPTEGESTDSEEETSATSGHPAELSPRPKQPLLSSGLYSSGSHSSSHERCSLSQSSTAQSLEDPPPFVEPSSEHPLSHKPEDTHTI KQKLALRLSERKKVIDEQAFLSPGSKGSTESGYFSRSESAEQQVSPPNTNAKSYAEIIFGKCGRIGQRTAMLTATSTQPLLPLSTEDKPS LVPLSVPRTQVIEHITKLITINEAVVDTSEIDSVKPRRSSLSRRSSMESPKSSLYREPLSSHSEKTKPEQSLLSLQHPPSTAPPVPLLRS HSMPSAACTISTPHHPFRGSYSFDDHITDSEALSHSSHVFTSHPRMLKRQPAIELPLGGEYSSEEPGPSSKDTASKPSDEVEPKESELTK KTKKGLKTKGVIYECNICGARYKKRDNYEAHKKYYCSELQIAKPISAGTHTSPEAEKSQIEHEPWSQMMHYKLGTTLELTPLRKRRKEKS LGDEEEPPAFESTKSQFGSPGPSDAARNLPLESTKSPAEPSKSVPSLEGPTGFQPRTPKPGSGSESGKERRTTSKEISVIQHTSSFEKSD SLEQPSGLEGEDKPLAQFPSPPPAPHGRSAHSLQPKLVRQPNIQVPEILVTEEPDRPDTEPEPPPKEPEKTEEFQWPQRSQTLAQLPAEK LPPKKKRLRLAEMAQSSGESSFESSVPLSRSPSQESNVSLSGSSRSASFERDDHGKAEAPSPSSDMRPKPLGTHMLTVPSHHPHAREMRR SASEQSPNVSHSAHMTETRSKSFDYGSLSLTGPSAPAPVAPPARVAPPERRKCFLVRQASLSRPPESELEVAPKGRQESEEPQPSSSKPS AKSSLSQISSAATSHGGPPGGKGPGQDRPPLGPTVPYTEALQVFHHPVAQTPLHEKPYLPPPVSLFSFQHLVQHEPGQSPEFFSTQAMSS LLSSPYSMPPLPPSLFQAPPLPLQPTVLHPGQLHLPQLMPHPANIPFRQPPSFLPMPYPTSSALSSGFFLPLQSQFALQLPGDVESHLPQ IKTSLAPLATGSAGLSPSTEYSSDIRLPPVAPPASSSAPTSAPPLALPACPDTMVSLVVPVRVQTNMPSYGSAMYTTLSQILVTQSQGSS ATVALPKFEEPPSKGTTVCGADVHEVGPGPSGLSEEQSRAFPTPYLRVPVTLPERKGTSLSSESILSLEGSSSTAGGSKRVLSPAGSLEL TMETQQQKRVKEEEASKADEKLELVKPCSVVLTSTEDGKRPEKSHLGNQGQGRRELEMLSSLSSDPSDTKEIPPLPHPALSHGTAPGSEA LKEYPQPSGKPHRRGLTPLSVKKEDSKEQPDLPSLAPPSSLPLSETSSRPAKSQEGTDSKKVLQFPSLHTTTNVSWCYLNYIKPNHIQHA DRRSSVYAGWCISLYNPNLPGVSTKAALSLLRSKQKVSKETYTMATAPHPEAGRLVPSSSRKPRMTEVHLPSLVSPEGQKDLARVEKEEE -------------------------------------------------------------- >36558_36558_2_HIVEP3-LRP8_HIVEP3_chr1_42041214_ENST00000372583_LRP8_chr1_53792664_ENST00000371454_length(amino acids)=1764AA_BP=1736 MDPEQSVKGTKKAEGSPRKRLTKGEAIQTSVSSSVPYPGSGTAATQESPAQELLAPQPFPGPSSVLREGSQEKTGQQQKPPKRPPIEASV HISQLPQHPLTPAFMSPGKPEHLLEGSTWQLVDPMRPGPSGSFVAPGLHPQSQLLPSHASIIPPEDLPGVPKVFVPRPSQVSLKPTEEAH KKERKPQKPGKYICQYCSRPCAKPSVLQKHIRSHTGERPYPCGPCGFSFKTKSNLYKHRKSHAHRIKAGLASGMGGEMYPHGLEMERIPG EEFEEPTEGESTDSEEETSATSGHPAELSPRPKQPLLSSGLYSSGSHSSSHERCSLSQSSTAQSLEDPPPFVEPSSEHPLSHKPEDTHTI KQKLALRLSERKKVIDEQAFLSPGSKGSTESGYFSRSESAEQQVSPPNTNAKSYAEIIFGKCGRIGQRTAMLTATSTQPLLPLSTEDKPS LVPLSVPRTQVIEHITKLITINEAVVDTSEIDSVKPRRSSLSRRSSMESPKSSLYREPLSSHSEKTKPEQSLLSLQHPPSTAPPVPLLRS HSMPSAACTISTPHHPFRGSYSFDDHITDSEALSHSSHVFTSHPRMLKRQPAIELPLGGEYSSEEPGPSSKDTASKPSDEVEPKESELTK KTKKGLKTKGVIYECNICGARYKKRDNYEAHKKYYCSELQIAKPISAGTHTSPEAEKSQIEHEPWSQMMHYKLGTTLELTPLRKRRKEKS LGDEEEPPAFESTKSQFGSPGPSDAARNLPLESTKSPAEPSKSVPSLEGPTGFQPRTPKPGSGSESGKERRTTSKEISVIQHTSSFEKSD SLEQPSGLEGEDKPLAQFPSPPPAPHGRSAHSLQPKLVRQPNIQVPEILVTEEPDRPDTEPEPPPKEPEKTEEFQWPQRSQTLAQLPAEK LPPKKKRLRLAEMAQSSGESSFESSVPLSRSPSQESNVSLSGSSRSASFERDDHGKAEAPSPSSDMRPKPLGTHMLTVPSHHPHAREMRR SASEQSPNVSHSAHMTETRSKSFDYGSLSLTGPSAPAPVAPPARVAPPERRKCFLVRQASLSRPPESELEVAPKGRQESEEPQPSSSKPS AKSSLSQISSAATSHGGPPGGKGPGQDRPPLGPTVPYTEALQVFHHPVAQTPLHEKPYLPPPVSLFSFQHLVQHEPGQSPEFFSTQAMSS LLSSPYSMPPLPPSLFQAPPLPLQPTVLHPGQLHLPQLMPHPANIPFRQPPSFLPMPYPTSSALSSGFFLPLQSQFALQLPGDVESHLPQ IKTSLAPLATGSAGLSPSTEYSSDIRLPPVAPPASSSAPTSAPPLALPACPDTMVSLVVPVRVQTNMPSYGSAMYTTLSQILVTQSQGSS ATVALPKFEEPPSKGTTVCGADVHEVGPGPSGLSEEQSRAFPTPYLRVPVTLPERKGTSLSSESILSLEGSSSTAGGSKRVLSPAGSLEL TMETQQQKRVKEEEASKADEKLELVKPCSVVLTSTEDGKRPEKSHLGNQGQGRRELEMLSSLSSDPSDTKEIPPLPHPALSHGTAPGSEA LKEYPQPSGKPHRRGLTPLSVKKEDSKEQPDLPSLAPPSSLPLSETSSRPAKSQEGTDSKKVLQFPSLHTTTNVSWCYLNYIKPNHIQHA DRRSSVYAGWCISLYNPNLPGVSTKAALSLLRSKQKVSKETYTMATAPHPEAGRLVPSSSRKPRMTEVHLPSLVSPEGQKDLARVEKEEE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:42041214/chr1:53792664) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HIVEP3 | LRP8 |
| FUNCTION: Plays a role of transcription factor; binds to recognition signal sequences (Rss heptamer) for somatic recombination of immunoglobulin and T-cell receptor gene segments; Binds also to the kappa-B motif of gene such as S100A4, involved in cell progression and differentiation. Kappa-B motif is a gene regulatory element found in promoters and enhancers of genes involved in immunity, inflammation, and growth and that responds to viral antigens, mitogens, and cytokines. Involvement of HIVEP3 in cell growth is strengthened by the fact that its down-regulation promotes cell cycle progression with ultimate formation of multinucleated giant cells. Strongly inhibits TNF-alpha-induced NF-kappa-B activation; Interferes with nuclear factor NF-kappa-B by several mechanisms: as transcription factor, by competing for Kappa-B motif and by repressing transcription in the nucleus; through a non transcriptional process, by inhibiting nuclear translocation of RELA by association with TRAF2, an adapter molecule in the tumor necrosis factor signaling, which blocks the formation of IKK complex. Interaction with TRAF proteins inhibits both NF-Kappa-B-mediated and c-Jun N-terminal kinase/JNK-mediated responses that include apoptosis and proinflammatory cytokine gene expression. Positively regulates the expression of IL2 in T-cell. Essential regulator of adult bone formation. {ECO:0000269|PubMed:11161801}. | FUNCTION: Cell surface receptor for Reelin (RELN) and apolipoprotein E (apoE)-containing ligands. LRP8 participates in transmitting the extracellular Reelin signal to intracellular signaling processes, by binding to DAB1 on its cytoplasmic tail. Reelin acts via both the VLDL receptor (VLDLR) and LRP8 to regulate DAB1 tyrosine phosphorylation and microtubule function in neurons. LRP8 has higher affinity for Reelin than VLDLR. LRP8 is thus a key component of the Reelin pathway which governs neuronal layering of the forebrain during embryonic brain development. Binds the endoplasmic reticulum resident receptor-associated protein (RAP). Binds dimers of beta 2-glycoprotein I and may be involved in the suppression of platelet aggregation in the vasculature. Highly expressed in the initial segment of the epididymis, where it affects the functional expression of clusterin and phospholipid hydroperoxide glutathione peroxidase (PHGPx), two proteins required for sperm maturation. May also function as an endocytic receptor. Not required for endocytic uptake of SEPP1 in the kidney which is mediated by LRP2 (By similarity). Together with its ligand, apolipoprotein E (apoE), may indirectly play a role in the suppression of the innate immune response by controlling the survival of myeloid-derived suppressor cells (By similarity). {ECO:0000250|UniProtKB:Q924X6, ECO:0000269|PubMed:12807892, ECO:0000269|PubMed:12899622, ECO:0000269|PubMed:12950167}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1442_1466 | 1735.6666666666667 | 2407.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1442_1466 | 1735.6666666666667 | 2407.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1442_1466 | 1735.6666666666667 | 2406.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1442_1466 | 1735.6666666666667 | 2406.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 308_334 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 378_414 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 797_819 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 844_880 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Glu/Pro-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 916_948 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 308_334 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 378_414 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 797_819 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 844_880 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Glu/Pro-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 916_948 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 308_334 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 378_414 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 797_819 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 844_880 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Glu/Pro-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 916_948 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 308_334 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 378_414 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 797_819 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 844_880 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Glu/Pro-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 916_948 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 903_909 | 1735.6666666666667 | 2407.0 | Motif | Nuclear localization signal |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 903_909 | 1735.6666666666667 | 2407.0 | Motif | Nuclear localization signal |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 903_909 | 1735.6666666666667 | 2406.0 | Motif | Nuclear localization signal |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 903_909 | 1735.6666666666667 | 2406.0 | Motif | Nuclear localization signal |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 192_242 | 1735.6666666666667 | 2407.0 | Region | Note=ZAS1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 211_1074 | 1735.6666666666667 | 2407.0 | Region | No DNA binding activity or transactivation activity%2C but complete prevention of TRAF-dependent NF-Kappa-B activation%3B associates with TRAF2 and JUN |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 264_287 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 862_883 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 192_242 | 1735.6666666666667 | 2407.0 | Region | Note=ZAS1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 211_1074 | 1735.6666666666667 | 2407.0 | Region | No DNA binding activity or transactivation activity%2C but complete prevention of TRAF-dependent NF-Kappa-B activation%3B associates with TRAF2 and JUN |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 264_287 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 862_883 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 192_242 | 1735.6666666666667 | 2406.0 | Region | Note=ZAS1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 211_1074 | 1735.6666666666667 | 2406.0 | Region | No DNA binding activity or transactivation activity%2C but complete prevention of TRAF-dependent NF-Kappa-B activation%3B associates with TRAF2 and JUN |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 264_287 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 862_883 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 192_242 | 1735.6666666666667 | 2406.0 | Region | Note=ZAS1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 211_1074 | 1735.6666666666667 | 2406.0 | Region | No DNA binding activity or transactivation activity%2C but complete prevention of TRAF-dependent NF-Kappa-B activation%3B associates with TRAF2 and JUN |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 264_287 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 862_883 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 192_214 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 220_242 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 640_670 | 1735.6666666666667 | 2407.0 | Zinc finger | CCHC HIVEP-type |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 192_214 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 220_242 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 640_670 | 1735.6666666666667 | 2407.0 | Zinc finger | CCHC HIVEP-type |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 192_214 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 220_242 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 640_670 | 1735.6666666666667 | 2406.0 | Zinc finger | CCHC HIVEP-type |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 192_214 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 220_242 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 640_670 | 1735.6666666666667 | 2406.0 | Zinc finger | CCHC HIVEP-type |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 126_164 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 166_202 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 205_246 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 258_295 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 298_334 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 336_375 | 41.333333333333336 | 964.0 | Domain | EGF-like 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 376_415 | 41.333333333333336 | 964.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 46_82 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 85_123 | 41.333333333333336 | 964.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 126_164 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 166_202 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 205_246 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 258_295 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 298_334 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 336_375 | 41.333333333333336 | 794.0 | Domain | EGF-like 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 376_415 | 41.333333333333336 | 794.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 46_82 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 85_123 | 41.333333333333336 | 794.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 126_164 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 166_202 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 205_246 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 258_295 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 298_334 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 336_375 | 41.333333333333336 | 701.0 | Domain | EGF-like 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 376_415 | 41.333333333333336 | 701.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 46_82 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 85_123 | 41.333333333333336 | 701.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 126_164 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 166_202 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 205_246 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 258_295 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 298_334 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 336_375 | 41.333333333333336 | 905.0 | Domain | EGF-like 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 376_415 | 41.333333333333336 | 905.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 46_82 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 85_123 | 41.333333333333336 | 905.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 740_798 | 41.333333333333336 | 964.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 740_798 | 41.333333333333336 | 794.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 740_798 | 41.333333333333336 | 701.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 740_798 | 41.333333333333336 | 905.0 | Region | Note=Clustered O-linked oligosaccharides | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 462_508 | 41.333333333333336 | 964.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 509_551 | 41.333333333333336 | 964.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 552_595 | 41.333333333333336 | 964.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 596_639 | 41.333333333333336 | 964.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 640_681 | 41.333333333333336 | 964.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 462_508 | 41.333333333333336 | 794.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 509_551 | 41.333333333333336 | 794.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 552_595 | 41.333333333333336 | 794.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 596_639 | 41.333333333333336 | 794.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 640_681 | 41.333333333333336 | 794.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 462_508 | 41.333333333333336 | 701.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 509_551 | 41.333333333333336 | 701.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 552_595 | 41.333333333333336 | 701.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 596_639 | 41.333333333333336 | 701.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 640_681 | 41.333333333333336 | 701.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 462_508 | 41.333333333333336 | 905.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 509_551 | 41.333333333333336 | 905.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 552_595 | 41.333333333333336 | 905.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 596_639 | 41.333333333333336 | 905.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 640_681 | 41.333333333333336 | 905.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 42_826 | 41.333333333333336 | 964.0 | Topological domain | Extracellular | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 848_963 | 41.333333333333336 | 964.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 42_826 | 41.333333333333336 | 794.0 | Topological domain | Extracellular | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 848_963 | 41.333333333333336 | 794.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 42_826 | 41.333333333333336 | 701.0 | Topological domain | Extracellular | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 848_963 | 41.333333333333336 | 701.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 42_826 | 41.333333333333336 | 905.0 | Topological domain | Extracellular | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 848_963 | 41.333333333333336 | 905.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000306052 | 0 | 19 | 827_847 | 41.333333333333336 | 964.0 | Transmembrane | Helical | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000347547 | 0 | 17 | 827_847 | 41.333333333333336 | 794.0 | Transmembrane | Helical | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000354412 | 0 | 16 | 827_847 | 41.333333333333336 | 701.0 | Transmembrane | Helical | |
| Tgene | LRP8 | chr1:42041214 | chr1:53792664 | ENST00000371454 | 0 | 18 | 827_847 | 41.333333333333336 | 905.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1907_1936 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1907_1936 | 1735.6666666666667 | 2407.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1907_1936 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1907_1936 | 1735.6666666666667 | 2406.0 | Compositional bias | Note=Ser-rich |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1754_1806 | 1735.6666666666667 | 2407.0 | Region | Note=ZAS2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1817_1872 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 2053_2148 | 1735.6666666666667 | 2407.0 | Region | Note=6 X 4 AA tandem repeats of S-P-X-[RK] |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1754_1806 | 1735.6666666666667 | 2407.0 | Region | Note=ZAS2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1817_1872 | 1735.6666666666667 | 2407.0 | Region | Note=Acidic 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 2053_2148 | 1735.6666666666667 | 2407.0 | Region | Note=6 X 4 AA tandem repeats of S-P-X-[RK] |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1754_1806 | 1735.6666666666667 | 2406.0 | Region | Note=ZAS2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1817_1872 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 2053_2148 | 1735.6666666666667 | 2406.0 | Region | Note=6 X 4 AA tandem repeats of S-P-X-[RK] |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1754_1806 | 1735.6666666666667 | 2406.0 | Region | Note=ZAS2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1817_1872 | 1735.6666666666667 | 2406.0 | Region | Note=Acidic 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 2053_2148 | 1735.6666666666667 | 2406.0 | Region | Note=6 X 4 AA tandem repeats of S-P-X-[RK] |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1964_1967 | 1735.6666666666667 | 2407.0 | Repeat | Note=1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1970_1973 | 1735.6666666666667 | 2407.0 | Repeat | Note=2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1993_1996 | 1735.6666666666667 | 2407.0 | Repeat | Note=3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1998_2001 | 1735.6666666666667 | 2407.0 | Repeat | Note=4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 2067_2070 | 1735.6666666666667 | 2407.0 | Repeat | Note=5 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 2079_2082 | 1735.6666666666667 | 2407.0 | Repeat | Note=6 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1964_1967 | 1735.6666666666667 | 2407.0 | Repeat | Note=1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1970_1973 | 1735.6666666666667 | 2407.0 | Repeat | Note=2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1993_1996 | 1735.6666666666667 | 2407.0 | Repeat | Note=3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1998_2001 | 1735.6666666666667 | 2407.0 | Repeat | Note=4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 2067_2070 | 1735.6666666666667 | 2407.0 | Repeat | Note=5 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 2079_2082 | 1735.6666666666667 | 2407.0 | Repeat | Note=6 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1964_1967 | 1735.6666666666667 | 2406.0 | Repeat | Note=1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1970_1973 | 1735.6666666666667 | 2406.0 | Repeat | Note=2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1993_1996 | 1735.6666666666667 | 2406.0 | Repeat | Note=3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1998_2001 | 1735.6666666666667 | 2406.0 | Repeat | Note=4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 2067_2070 | 1735.6666666666667 | 2406.0 | Repeat | Note=5 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 2079_2082 | 1735.6666666666667 | 2406.0 | Repeat | Note=6 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1964_1967 | 1735.6666666666667 | 2406.0 | Repeat | Note=1 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1970_1973 | 1735.6666666666667 | 2406.0 | Repeat | Note=2 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1993_1996 | 1735.6666666666667 | 2406.0 | Repeat | Note=3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1998_2001 | 1735.6666666666667 | 2406.0 | Repeat | Note=4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 2067_2070 | 1735.6666666666667 | 2406.0 | Repeat | Note=5 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 2079_2082 | 1735.6666666666667 | 2406.0 | Repeat | Note=6 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1754_1776 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000247584 | - | 4 | 8 | 1782_1806 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1754_1776 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372583 | - | 5 | 9 | 1782_1806 | 1735.6666666666667 | 2407.0 | Zinc finger | C2H2-type 4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1754_1776 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000372584 | - | 4 | 8 | 1782_1806 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 4 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1754_1776 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 3 |
| Hgene | HIVEP3 | chr1:42041214 | chr1:53792664 | ENST00000429157 | - | 4 | 8 | 1782_1806 | 1735.6666666666667 | 2406.0 | Zinc finger | C2H2-type 4 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HIVEP3 | |
| LRP8 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HIVEP3-LRP8 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HIVEP3-LRP8 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

