| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HNRNPUL1-BRSK1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HNRNPUL1-BRSK1 | FusionPDB ID: 37305 | FusionGDB2.0 ID: 37305 | Hgene | Tgene | Gene symbol | HNRNPUL1 | BRSK1 | Gene ID | 11100 | 84446 |
| Gene name | heterogeneous nuclear ribonucleoprotein U like 1 | BR serine/threonine kinase 1 | |
| Synonyms | E1B-AP5|E1BAP5|HNRPUL1 | hSAD1 | |
| Cytomap | 19q13.2 | 19q13.42 | |
| Type of gene | protein-coding | protein-coding | |
| Description | heterogeneous nuclear ribonucleoprotein U-like protein 1E1B 55kDa associated protein 5E1B-55 kDa-associated protein 5adenovirus early region 1B-associated protein 5 | serine/threonine-protein kinase BRSK1BR serine/threonine-protein kinase 1SAD1 homologSAD1 kinaseSadB kinase short isoformbrain-selective kinase 1brain-specific serine/threonine-protein kinase 1protein kinase SAD1Aserine/threonine-protein kinase SA | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q9BUJ2 | Q8TDC3 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000263367, ENST00000352456, ENST00000392006, ENST00000593587, ENST00000595018, ENST00000602130, ENST00000378215, ENST00000594207, | ENST00000326848, ENST00000588584, ENST00000309383, ENST00000585418, ENST00000590333, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 18 X 14 X 12=3024 | 5 X 5 X 4=100 |
| # samples | 20 | 5 | |
| ** MAII score | log2(20/3024*10)=-3.91838623444635 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HNRNPUL1 [Title/Abstract] AND BRSK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HNRNPUL1(41787180)-BRSK1(55795889), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HNRNPUL1-BRSK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. HNRNPUL1-BRSK1 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BRSK1 | GO:0000086 | G2/M transition of mitotic cell cycle | 15150265 |
| Tgene | BRSK1 | GO:0006468 | protein phosphorylation | 15150265 |
| Tgene | BRSK1 | GO:0006974 | cellular response to DNA damage stimulus | 15150265 |
| Tgene | BRSK1 | GO:0007095 | mitotic G2 DNA damage checkpoint | 15150265 |
| Tgene | BRSK1 | GO:0009411 | response to UV | 15150265 |
| Tgene | BRSK1 | GO:0010212 | response to ionizing radiation | 15150265 |
| Tgene | BRSK1 | GO:0018105 | peptidyl-serine phosphorylation | 15150265 |
 Fusion gene breakpoints across HNRNPUL1 (5'-gene) Fusion gene breakpoints across HNRNPUL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
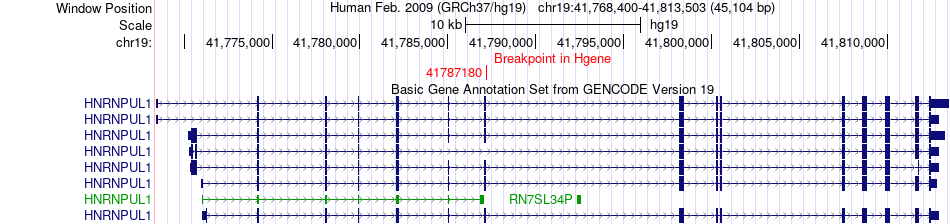 |
 Fusion gene breakpoints across BRSK1 (3'-gene) Fusion gene breakpoints across BRSK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
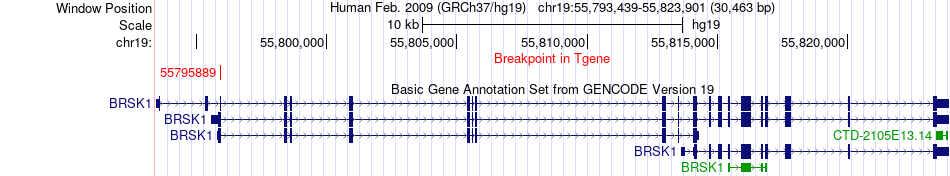 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-C5-A907-01A | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000352456 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3515 | 791 | 11 | 3049 | 1012 |
| ENST00000595018 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3492 | 768 | 45 | 3026 | 993 |
| ENST00000392006 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3896 | 1172 | 44 | 3430 | 1128 |
| ENST00000602130 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3776 | 1052 | 53 | 3310 | 1085 |
| ENST00000593587 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3517 | 793 | 94 | 3051 | 985 |
| ENST00000263367 | HNRNPUL1 | chr19 | 41787180 | + | ENST00000590333 | BRSK1 | chr19 | 55795889 | + | 3650 | 926 | 161 | 3184 | 1007 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000352456 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.002145738 | 0.9978543 |
| ENST00000595018 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.002256464 | 0.9977436 |
| ENST00000392006 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.003378014 | 0.99662197 |
| ENST00000602130 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.003245843 | 0.9967541 |
| ENST00000593587 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.002510866 | 0.9974891 |
| ENST00000263367 | ENST00000590333 | HNRNPUL1 | chr19 | 41787180 | + | BRSK1 | chr19 | 55795889 | + | 0.00237696 | 0.997623 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >37305_37305_1_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000263367_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=1007AA_BP=255 MRKEDPGICRWMRSWAFSYLDAMDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAYRPEMKTEMKQGAPTSFLPPEASQLKPDRQQF QSRKRPYEENRGRGYFEHREDRRGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARDRSSGYPLTIEGFAYLWSGARASYGVRRGRVC FEMKINEEISVKHLPSTEPDPHVVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFENYGDKFAENDVIGCFAHAQYVGPYRLEKTLG KGQTGLVKLGVHCITGQKVAIKIVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPK EARKFFRQIVSALDFCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGRRADMWSCGVIL FALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAMRSLPS NGELDPDVLESMASLGCFRDRERLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLPPRNDVDPPRKRVDSPMLSRHGKRRPERKSME VLSITDAGGGGSPVPTRRALEMAQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPGAGDEARGGGSPTSKTQTLPSRGPRGGGAGEQ PPPPSARSTPLPGPPGSPRSSGGTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWRSRLNSIRNSFLGSPRFHRRKMQVPTAEEMSS LTPESSPELAKRSWFGNFISLDKEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLSQTSFRAEYKASGGPSVFQKPVRFQVDISSSE GPEPSPRRDGSGGGGIYSVTFTLISGPSRRFKRVVETIQAQLLSTHDQPSVQALADEKNGAQTRPAGAPPRSLQPPPGRPDPELSSSPRR -------------------------------------------------------------- >37305_37305_2_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000352456_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=1012AA_BP=260 MGGERERGSRTWARGGVSGLEPLGPHAMDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAYRPEMKTEMKQGAPTSFLPPEASQLKP DRQQFQSRKRPYEENRGRGYFEHREDRRGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARDRSSGYPLTIEGFAYLWSGARASYGVR RGRVCFEMKINEEISVKHLPSTEPDPHVVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFENYGDKFAENDVIGCFAHAQYVGPYRL EKTLGKGQTGLVKLGVHCITGQKVAIKIVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKG RLTPKEARKFFRQIVSALDFCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGRRADMWS CGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAM RSLPSNGELDPDVLESMASLGCFRDRERLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLPPRNDVDPPRKRVDSPMLSRHGKRRPE RKSMEVLSITDAGGGGSPVPTRRALEMAQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPGAGDEARGGGSPTSKTQTLPSRGPRGG GAGEQPPPPSARSTPLPGPPGSPRSSGGTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWRSRLNSIRNSFLGSPRFHRRKMQVPTA EEMSSLTPESSPELAKRSWFGNFISLDKEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLSQTSFRAEYKASGGPSVFQKPVRFQVD ISSSEGPEPSPRRDGSGGGGIYSVTFTLISGPSRRFKRVVETIQAQLLSTHDQPSVQALADEKNGAQTRPAGAPPRSLQPPPGRPDPELS -------------------------------------------------------------- >37305_37305_3_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000392006_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=1128AA_BP=376 MSRCRHWSGPPPFPPSPPDRKGLRGTEPWEAGPGSGATPGARAMDVRRLKVNELREELQRRGLDTRGLKAELAERLQAALEAEEPDDERE LDADDEPGRPGHINEEVETEGGSELEGTAQPPPPGLQPHAEPGGYSGPDGHYAMDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAY RPEMKTEMKQGAPTSFLPPEASQLKPDRQQFQSRKRPYEENRGRGYFEHREDRRGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARD RSSGYPLTIEGFAYLWSGARASYGVRRGRVCFEMKINEEISVKHLPSTEPDPHVVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFE NYGDKFAENDVIGCFAHAQYVGPYRLEKTLGKGQTGLVKLGVHCITGQKVAIKIVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVY ENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSALDFCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETS CGSPHYACPEVIKGEKYDGRRADMWSCGVILFALLVGALPFDDDNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQ KHPWYLGGKHEPDPCLEPAPGRRVAMRSLPSNGELDPDVLESMASLGCFRDRERLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLP PRNDVDPPRKRVDSPMLSRHGKRRPERKSMEVLSITDAGGGGSPVPTRRALEMAQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPG AGDEARGGGSPTSKTQTLPSRGPRGGGAGEQPPPPSARSTPLPGPPGSPRSSGGTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWR SRLNSIRNSFLGSPRFHRRKMQVPTAEEMSSLTPESSPELAKRSWFGNFISLDKEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLS QTSFRAEYKASGGPSVFQKPVRFQVDISSSEGPEPSPRRDGSGGGGIYSVTFTLISGPSRRFKRVVETIQAQLLSTHDQPSVQALADEKN -------------------------------------------------------------- >37305_37305_4_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000593587_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=985AA_BP=233 MDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAYRPEMKTEMKQGAPTSFLPPEASQLKPDRQQFQSRKRPYEENRGRGYFEHREDR RGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARDRSSGYPLTIEGFAYLWSGARASYGVRRGRVCFEMKINEEISVKHLPSTEPDPH VVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFENYGDKFAENDVIGCFAHAQYVGPYRLEKTLGKGQTGLVKLGVHCITGQKVAIK IVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSALDFCHSYSIC HRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGRRADMWSCGVILFALLVGALPFDDDNLRQLLEKV KRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAMRSLPSNGELDPDVLESMASLGCFRDRE RLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLPPRNDVDPPRKRVDSPMLSRHGKRRPERKSMEVLSITDAGGGGSPVPTRRALEM AQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPGAGDEARGGGSPTSKTQTLPSRGPRGGGAGEQPPPPSARSTPLPGPPGSPRSSG GTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWRSRLNSIRNSFLGSPRFHRRKMQVPTAEEMSSLTPESSPELAKRSWFGNFISLD KEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLSQTSFRAEYKASGGPSVFQKPVRFQVDISSSEGPEPSPRRDGSGGGGIYSVTFT -------------------------------------------------------------- >37305_37305_5_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000595018_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=993AA_BP=241 MEPLGPHAMDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAYRPEMKTEMKQGAPTSFLPPEASQLKPDRQQFQSRKRPYEENRGRG YFEHREDRRGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARDRSSGYPLTIEGFAYLWSGARASYGVRRGRVCFEMKINEEISVKHL PSTEPDPHVVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFENYGDKFAENDVIGCFAHAQYVGPYRLEKTLGKGQTGLVKLGVHCI TGQKVAIKIVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSALD FCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGRRADMWSCGVILFALLVGALPFDDDN LRQLLEKVKRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAMRSLPSNGELDPDVLESMAS LGCFRDRERLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLPPRNDVDPPRKRVDSPMLSRHGKRRPERKSMEVLSITDAGGGGSPV PTRRALEMAQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPGAGDEARGGGSPTSKTQTLPSRGPRGGGAGEQPPPPSARSTPLPGP PGSPRSSGGTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWRSRLNSIRNSFLGSPRFHRRKMQVPTAEEMSSLTPESSPELAKRSW FGNFISLDKEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLSQTSFRAEYKASGGPSVFQKPVRFQVDISSSEGPEPSPRRDGSGGG GIYSVTFTLISGPSRRFKRVVETIQAQLLSTHDQPSVQALADEKNGAQTRPAGAPPRSLQPPPGRPDPELSSSPRRGPPKDKKLLATNGT -------------------------------------------------------------- >37305_37305_6_HNRNPUL1-BRSK1_HNRNPUL1_chr19_41787180_ENST00000602130_BRSK1_chr19_55795889_ENST00000590333_length(amino acids)=1085AA_BP=333 MDVRRLKVNELREELQRRGLDTRGLKAELAERLQAALEAEEPDDERELDADDEPGRPGHINEEVETEGGSELEGTAQPPPPGLQPHAEPG GYSGPDGHYAMDNITRQNQFYDTQVIKQENESGYERRPLEMEQQQAYRPEMKTEMKQGAPTSFLPPEASQLKPDRQQFQSRKRPYEENRG RGYFEHREDRRGRSPQPPAEEDEDDFDDTLVAIDTYNCDLHFKVARDRSSGYPLTIEGFAYLWSGARASYGVRRGRVCFEMKINEEISVK HLPSTEPDPHVVRIGWSLDSCSTQLGEEPFSYGYGGTGKKSTNSRFENYGDKFAENDVIGCFAHAQYVGPYRLEKTLGKGQTGLVKLGVH CITGQKVAIKIVNREKLSESVLMKVEREIAILKLIEHPHVLKLHDVYENKKYLYLVLEHVSGGELFDYLVKKGRLTPKEARKFFRQIVSA LDFCHSYSICHRDLKPENLLLDEKNNIRIADFGMASLQVGDSLLETSCGSPHYACPEVIKGEKYDGRRADMWSCGVILFALLVGALPFDD DNLRQLLEKVKRGVFHMPHFIPPDCQSLLRGMIEVEPEKRLSLEQIQKHPWYLGGKHEPDPCLEPAPGRRVAMRSLPSNGELDPDVLESM ASLGCFRDRERLHRELRSEEENQEKMIYYLLLDRKERYPSCEDQDLPPRNDVDPPRKRVDSPMLSRHGKRRPERKSMEVLSITDAGGGGS PVPTRRALEMAQHSQRSRSVSGASTGLSSSPLSSPRSPVFSFSPEPGAGDEARGGGSPTSKTQTLPSRGPRGGGAGEQPPPPSARSTPLP GPPGSPRSSGGTPLHSPLHTPRASPTGTPGTTPPPSPGGGVGGAAWRSRLNSIRNSFLGSPRFHRRKMQVPTAEEMSSLTPESSPELAKR SWFGNFISLDKEEQIFLVLKDKPLSSIKADIVHAFLSIPSLSHSVLSQTSFRAEYKASGGPSVFQKPVRFQVDISSSEGPEPSPRRDGSG GGGIYSVTFTLISGPSRRFKRVVETIQAQLLSTHDQPSVQALADEKNGAQTRPAGAPPRSLQPPPGRPDPELSSSPRRGPPKDKKLLATN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:41787180/chr19:55795889) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HNRNPUL1 | BRSK1 |
| FUNCTION: Acts as a basic transcriptional regulator. Represses basic transcription driven by several virus and cellular promoters. When associated with BRD7, activates transcription of glucocorticoid-responsive promoter in the absence of ligand-stimulation. Plays also a role in mRNA processing and transport. Binds avidly to poly(G) and poly(C) RNA homopolymers in vitro. {ECO:0000269|PubMed:12489984, ECO:0000269|PubMed:9733834}. | FUNCTION: Serine/threonine-protein kinase that plays a key role in polarization of neurons and centrosome duplication. Phosphorylates CDC25B, CDC25C, MAPT/TAU, RIMS1, TUBG1, TUBG2 and WEE1. Following phosphorylation and activation by STK11/LKB1, acts as a key regulator of polarization of cortical neurons, probably by mediating phosphorylation of microtubule-associated proteins such as MAPT/TAU at 'Thr-529' and 'Ser-579'. Also regulates neuron polarization by mediating phosphorylation of WEE1 at 'Ser-642' in postmitotic neurons, leading to down-regulate WEE1 activity in polarized neurons. In neurons, localizes to synaptic vesicles and plays a role in neurotransmitter release, possibly by phosphorylating RIMS1. Also acts as a positive regulator of centrosome duplication by mediating phosphorylation of gamma-tubulin (TUBG1 and TUBG2) at 'Ser-131', leading to translocation of gamma-tubulin and its associated proteins to the centrosome. Involved in the UV-induced DNA damage checkpoint response, probably by inhibiting CDK1 activity through phosphorylation and activation of WEE1, and inhibition of CDC25B and CDC25C. {ECO:0000269|PubMed:14976552, ECO:0000269|PubMed:15150265, ECO:0000269|PubMed:20026642, ECO:0000269|PubMed:21985311}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 3_37 | 333.0 | 857.0 | Domain | SAP |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 3_37 | 233.0 | 757.0 | Domain | SAP |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 3_37 | 233.0 | 757.0 | Domain | SAP |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 3_37 | 333.0 | 805.0 | Domain | SAP |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000309383 | 0 | 19 | 492_540 | 0 | 779.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000585418 | 0 | 10 | 492_540 | 0 | 344.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000590333 | 1 | 21 | 492_540 | 42.0 | 795.0 | Compositional bias | Note=Pro-rich | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000309383 | 0 | 19 | 314_356 | 0 | 779.0 | Domain | UBA | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000309383 | 0 | 19 | 34_285 | 0 | 779.0 | Domain | Protein kinase | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000585418 | 0 | 10 | 314_356 | 0 | 344.0 | Domain | UBA | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000585418 | 0 | 10 | 34_285 | 0 | 344.0 | Domain | Protein kinase | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000590333 | 1 | 21 | 314_356 | 42.0 | 795.0 | Domain | UBA | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000309383 | 0 | 19 | 40_48 | 0 | 779.0 | Nucleotide binding | ATP | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000585418 | 0 | 10 | 40_48 | 0 | 344.0 | Nucleotide binding | ATP | |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000590333 | 1 | 21 | 40_48 | 42.0 | 795.0 | Nucleotide binding | ATP |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 613_666 | 0 | 753.0 | Compositional bias | Note=Gly-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 670_689 | 0 | 753.0 | Compositional bias | Note=Asn-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 692_811 | 0 | 753.0 | Compositional bias | Note=Pro-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 757_845 | 0 | 753.0 | Compositional bias | Note=Tyr-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 806_832 | 0 | 753.0 | Compositional bias | Note=Gln-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 613_666 | 333.0 | 857.0 | Compositional bias | Note=Gly-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 670_689 | 333.0 | 857.0 | Compositional bias | Note=Asn-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 692_811 | 333.0 | 857.0 | Compositional bias | Note=Pro-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 757_845 | 333.0 | 857.0 | Compositional bias | Note=Tyr-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 806_832 | 333.0 | 857.0 | Compositional bias | Note=Gln-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 613_666 | 233.0 | 757.0 | Compositional bias | Note=Gly-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 670_689 | 233.0 | 757.0 | Compositional bias | Note=Asn-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 692_811 | 233.0 | 757.0 | Compositional bias | Note=Pro-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 757_845 | 233.0 | 757.0 | Compositional bias | Note=Tyr-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 806_832 | 233.0 | 757.0 | Compositional bias | Note=Gln-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 613_666 | 233.0 | 757.0 | Compositional bias | Note=Gly-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 670_689 | 233.0 | 757.0 | Compositional bias | Note=Asn-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 692_811 | 233.0 | 757.0 | Compositional bias | Note=Pro-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 757_845 | 233.0 | 757.0 | Compositional bias | Note=Tyr-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 806_832 | 233.0 | 757.0 | Compositional bias | Note=Gln-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 613_666 | 333.0 | 805.0 | Compositional bias | Note=Gly-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 670_689 | 333.0 | 805.0 | Compositional bias | Note=Asn-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 692_811 | 333.0 | 805.0 | Compositional bias | Note=Pro-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 757_845 | 333.0 | 805.0 | Compositional bias | Note=Tyr-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 806_832 | 333.0 | 805.0 | Compositional bias | Note=Gln-rich |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 191_388 | 0 | 753.0 | Domain | B30.2/SPRY |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 3_37 | 0 | 753.0 | Domain | SAP |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 191_388 | 333.0 | 857.0 | Domain | B30.2/SPRY |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 191_388 | 233.0 | 757.0 | Domain | B30.2/SPRY |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 191_388 | 233.0 | 757.0 | Domain | B30.2/SPRY |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 191_388 | 333.0 | 805.0 | Domain | B30.2/SPRY |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 612_658 | 0 | 753.0 | Region | Note=Necessary for transcription repression |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 612_658 | 333.0 | 857.0 | Region | Note=Necessary for transcription repression |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 612_658 | 233.0 | 757.0 | Region | Note=Necessary for transcription repression |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 612_658 | 233.0 | 757.0 | Region | Note=Necessary for transcription repression |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 612_658 | 333.0 | 805.0 | Region | Note=Necessary for transcription repression |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 612_614 | 0 | 753.0 | Repeat | Note=1-1 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 620_622 | 0 | 753.0 | Repeat | Note=1-2 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 639_641 | 0 | 753.0 | Repeat | Note=1-3 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 645_647 | 0 | 753.0 | Repeat | Note=1-4 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000378215 | + | 1 | 14 | 656_658 | 0 | 753.0 | Repeat | Note=1-5 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 612_614 | 333.0 | 857.0 | Repeat | Note=1-1 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 620_622 | 333.0 | 857.0 | Repeat | Note=1-2 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 639_641 | 333.0 | 857.0 | Repeat | Note=1-3 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 645_647 | 333.0 | 857.0 | Repeat | Note=1-4 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000392006 | + | 7 | 15 | 656_658 | 333.0 | 857.0 | Repeat | Note=1-5 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 612_614 | 233.0 | 757.0 | Repeat | Note=1-1 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 620_622 | 233.0 | 757.0 | Repeat | Note=1-2 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 639_641 | 233.0 | 757.0 | Repeat | Note=1-3 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 645_647 | 233.0 | 757.0 | Repeat | Note=1-4 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000593587 | + | 7 | 15 | 656_658 | 233.0 | 757.0 | Repeat | Note=1-5 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 612_614 | 233.0 | 757.0 | Repeat | Note=1-1 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 620_622 | 233.0 | 757.0 | Repeat | Note=1-2 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 639_641 | 233.0 | 757.0 | Repeat | Note=1-3 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 645_647 | 233.0 | 757.0 | Repeat | Note=1-4 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000595018 | + | 7 | 15 | 656_658 | 233.0 | 757.0 | Repeat | Note=1-5 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 612_614 | 333.0 | 805.0 | Repeat | Note=1-1 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 620_622 | 333.0 | 805.0 | Repeat | Note=1-2 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 639_641 | 333.0 | 805.0 | Repeat | Note=1-3 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 645_647 | 333.0 | 805.0 | Repeat | Note=1-4 |
| Hgene | HNRNPUL1 | chr19:41787180 | chr19:55795889 | ENST00000602130 | + | 7 | 15 | 656_658 | 333.0 | 805.0 | Repeat | Note=1-5 |
| Tgene | BRSK1 | chr19:41787180 | chr19:55795889 | ENST00000590333 | 1 | 21 | 34_285 | 42.0 | 795.0 | Domain | Protein kinase |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HNRNPUL1 | |
| BRSK1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HNRNPUL1-BRSK1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HNRNPUL1-BRSK1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

