| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HOOK3-IKBKB |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HOOK3-IKBKB | FusionPDB ID: 37391 | FusionGDB2.0 ID: 37391 | Hgene | Tgene | Gene symbol | HOOK3 | IKBKB | Gene ID | 84376 | 3551 |
| Gene name | hook microtubule tethering protein 3 | inhibitor of nuclear factor kappa B kinase subunit beta | |
| Synonyms | HK3 | IKK-beta|IKK2|IKKB|IMD15|IMD15A|IMD15B|NFKBIKB | |
| Cytomap | 8p11.21 | 8p11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein Hook homolog 3h-hook3hHK3hook homolog 3 | inhibitor of nuclear factor kappa-B kinase subunit betaI-kappa-B kinase 2I-kappa-B-kinase betainhibitor of kappa light polypeptide gene enhancer in B-cells, kinase betanuclear factor NF-kappa-B inhibitor kinase beta | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q86VS8 | O14920 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000307602, ENST00000524839, | ENST00000379708, ENST00000518983, ENST00000522147, ENST00000522785, ENST00000416505, ENST00000519735, ENST00000520810, ENST00000520835, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 13 X 9=1872 | 11 X 14 X 4=616 |
| # samples | 19 | 14 | |
| ** MAII score | log2(19/1872*10)=-3.30050911125246 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/616*10)=-2.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HOOK3 [Title/Abstract] AND IKBKB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HOOK3(42798588)-IKBKB(42162705), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. HOOK3-IKBKB seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. HOOK3-IKBKB seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. HOOK3-IKBKB seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. HOOK3-IKBKB seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | IKBKB | GO:0006468 | protein phosphorylation | 15084260|20434986 |
| Tgene | IKBKB | GO:0018105 | peptidyl-serine phosphorylation | 21399639 |
| Tgene | IKBKB | GO:0042325 | regulation of phosphorylation | 26212789 |
| Tgene | IKBKB | GO:0045944 | positive regulation of transcription by RNA polymerase II | 23091055|23453807 |
| Tgene | IKBKB | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 15790681 |
| Tgene | IKBKB | GO:0071356 | cellular response to tumor necrosis factor | 23091055 |
 Fusion gene breakpoints across HOOK3 (5'-gene) Fusion gene breakpoints across HOOK3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
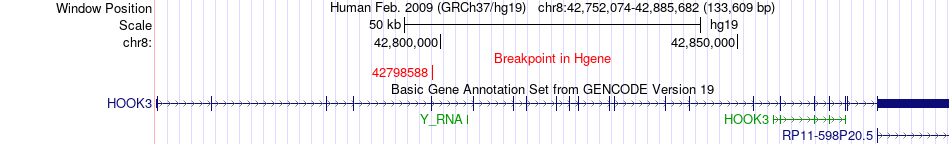 |
 Fusion gene breakpoints across IKBKB (3'-gene) Fusion gene breakpoints across IKBKB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
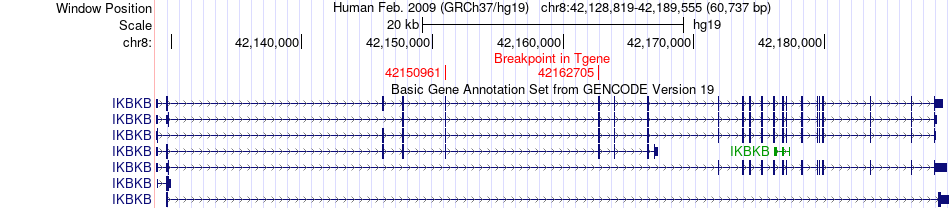 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A2-A0D1-01A | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42150961 | + |
| ChimerDB4 | BRCA | TCGA-A2-A0D1-01A | HOOK3 | chr8 | 42798588 | - | IKBKB | chr8 | 42162705 | + |
| ChimerDB4 | BRCA | TCGA-A2-A0D1-01A | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42162705 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000307602 | HOOK3 | chr8 | 42798588 | + | ENST00000520810 | IKBKB | chr8 | 42162705 | + | 3112 | 600 | 182 | 2482 | 766 |
| ENST00000307602 | HOOK3 | chr8 | 42798588 | + | ENST00000416505 | IKBKB | chr8 | 42162705 | + | 2636 | 600 | 182 | 2482 | 766 |
| ENST00000307602 | HOOK3 | chr8 | 42798588 | + | ENST00000519735 | IKBKB | chr8 | 42162705 | + | 1208 | 600 | 182 | 982 | 266 |
| ENST00000307602 | HOOK3 | chr8 | 42798588 | + | ENST00000520835 | IKBKB | chr8 | 42162705 | + | 2568 | 600 | 182 | 2482 | 766 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000307602 | ENST00000520810 | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42162705 | + | 0.00313604 | 0.99686396 |
| ENST00000307602 | ENST00000416505 | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42162705 | + | 0.005803003 | 0.994197 |
| ENST00000307602 | ENST00000519735 | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42162705 | + | 0.00492399 | 0.995076 |
| ENST00000307602 | ENST00000520835 | HOOK3 | chr8 | 42798588 | + | IKBKB | chr8 | 42162705 | + | 0.005431653 | 0.99456835 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >37391_37391_1_HOOK3-IKBKB_HOOK3_chr8_42798588_ENST00000307602_IKBKB_chr8_42162705_ENST00000416505_length(amino acids)=766AA_BP=139 MAAAGKMFSVESLERAELCESLLTWIQTFNVDAPCQTVEDLTNGVVMAQVLQKIDPAYFDENWLNRIKTEVGDNWRLKISNLKKILKGIL DYNHEILGQQINDFTLPDVNLIGEHSDAAELGRMLQLILGCAVNCEQKQASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYA KELDQGSLCTSFVGTLQYLAPELLEQQKYTVTVDYWSFGTLAFECITGFRPFLPNWQPVQWHSKVRQKSEVDIVVSEDLNGTVKFSSSLP YPNNLNSVLAERLEKWLQLMLMWHPRQRGTDPTYGPNGCFKALDDILNLKLVHILNMVTGTIHTYPVTEDESLQSLKARIQQDTGIPEED QELLQEAGLALIPDKPATQCISDGKLNEGHTLDMDLVFLFDNSKITYETQISPRPQPESVSCILQEPKRNLAFFQLRKVWGQVWHSIQTL KEDCNRLQQGQRAAMMNLLRNNSCLSKMKNSMASMSQQLKAKLDFFKTSIQIDLEKYSEQTEFGITSDKLLLAWREMEQAVELCGRENEV KLLVERMMALQTDIVDLQRSPMGRKQGGTLDDLEEQARELYRRLREKPRDQRTEGDSQEMVRLLLQAIQSFEKKVRVIYTQLSKTVVCKQ KALELLPKVEEVVSLMNEDEKTVVRLQEKRQKELWNLLKIACSKVRGPVSGSPDSMNASRLSQPGQLMSQPSTASNSLPEPAKKSEELVA -------------------------------------------------------------- >37391_37391_2_HOOK3-IKBKB_HOOK3_chr8_42798588_ENST00000307602_IKBKB_chr8_42162705_ENST00000519735_length(amino acids)=266AA_BP=139 MAAAGKMFSVESLERAELCESLLTWIQTFNVDAPCQTVEDLTNGVVMAQVLQKIDPAYFDENWLNRIKTEVGDNWRLKISNLKKILKGIL DYNHEILGQQINDFTLPDVNLIGEHSDAAELGRMLQLILGCAVNCEQKQASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYA -------------------------------------------------------------- >37391_37391_3_HOOK3-IKBKB_HOOK3_chr8_42798588_ENST00000307602_IKBKB_chr8_42162705_ENST00000520810_length(amino acids)=766AA_BP=139 MAAAGKMFSVESLERAELCESLLTWIQTFNVDAPCQTVEDLTNGVVMAQVLQKIDPAYFDENWLNRIKTEVGDNWRLKISNLKKILKGIL DYNHEILGQQINDFTLPDVNLIGEHSDAAELGRMLQLILGCAVNCEQKQASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYA KELDQGSLCTSFVGTLQYLAPELLEQQKYTVTVDYWSFGTLAFECITGFRPFLPNWQPVQWHSKVRQKSEVDIVVSEDLNGTVKFSSSLP YPNNLNSVLAERLEKWLQLMLMWHPRQRGTDPTYGPNGCFKALDDILNLKLVHILNMVTGTIHTYPVTEDESLQSLKARIQQDTGIPEED QELLQEAGLALIPDKPATQCISDGKLNEGHTLDMDLVFLFDNSKITYETQISPRPQPESVSCILQEPKRNLAFFQLRKVWGQVWHSIQTL KEDCNRLQQGQRAAMMNLLRNNSCLSKMKNSMASMSQQLKAKLDFFKTSIQIDLEKYSEQTEFGITSDKLLLAWREMEQAVELCGRENEV KLLVERMMALQTDIVDLQRSPMGRKQGGTLDDLEEQARELYRRLREKPRDQRTEGDSQEMVRLLLQAIQSFEKKVRVIYTQLSKTVVCKQ KALELLPKVEEVVSLMNEDEKTVVRLQEKRQKELWNLLKIACSKVRGPVSGSPDSMNASRLSQPGQLMSQPSTASNSLPEPAKKSEELVA -------------------------------------------------------------- >37391_37391_4_HOOK3-IKBKB_HOOK3_chr8_42798588_ENST00000307602_IKBKB_chr8_42162705_ENST00000520835_length(amino acids)=766AA_BP=139 MAAAGKMFSVESLERAELCESLLTWIQTFNVDAPCQTVEDLTNGVVMAQVLQKIDPAYFDENWLNRIKTEVGDNWRLKISNLKKILKGIL DYNHEILGQQINDFTLPDVNLIGEHSDAAELGRMLQLILGCAVNCEQKQASALRYLHENRIIHRDLKPENIVLQQGEQRLIHKIIDLGYA KELDQGSLCTSFVGTLQYLAPELLEQQKYTVTVDYWSFGTLAFECITGFRPFLPNWQPVQWHSKVRQKSEVDIVVSEDLNGTVKFSSSLP YPNNLNSVLAERLEKWLQLMLMWHPRQRGTDPTYGPNGCFKALDDILNLKLVHILNMVTGTIHTYPVTEDESLQSLKARIQQDTGIPEED QELLQEAGLALIPDKPATQCISDGKLNEGHTLDMDLVFLFDNSKITYETQISPRPQPESVSCILQEPKRNLAFFQLRKVWGQVWHSIQTL KEDCNRLQQGQRAAMMNLLRNNSCLSKMKNSMASMSQQLKAKLDFFKTSIQIDLEKYSEQTEFGITSDKLLLAWREMEQAVELCGRENEV KLLVERMMALQTDIVDLQRSPMGRKQGGTLDDLEEQARELYRRLREKPRDQRTEGDSQEMVRLLLQAIQSFEKKVRVIYTQLSKTVVCKQ KALELLPKVEEVVSLMNEDEKTVVRLQEKRQKELWNLLKIACSKVRGPVSGSPDSMNASRLSQPGQLMSQPSTASNSLPEPAKKSEELVA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:42798588/chr8:42162705) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HOOK3 | IKBKB |
| FUNCTION: Probably serves as a target for the spiC protein from Salmonella typhimurium, which inactivates it, leading to a strong alteration in cellular trafficking (By similarity). Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). May regulate clearance of endocytosed receptors such as MSR1. Participates in defining the architecture and localization of the Golgi complex. Acts as an adapter protein linking the dynein motor complex to various cargos and converts dynein from a non-processive to a highly processive motor in the presence of dynactin. Facilitates the interaction between dynein and dynactin and activates dynein processivity (the ability to move along a microtubule for a long distance without falling off the track) (PubMed:25035494). FHF complex promotes the distribution of AP-4 complex to the perinuclear area of the cell (PubMed:32073997). {ECO:0000250|UniProtKB:Q8BUK6, ECO:0000269|PubMed:11238449, ECO:0000269|PubMed:17237231, ECO:0000269|PubMed:18799622, ECO:0000269|PubMed:25035494, ECO:0000269|PubMed:32073997}.; FUNCTION: (Microbial infection) May serve as a target for the spiC protein from Salmonella typhimurium, which inactivates it, leading to a strong alteration in cellular trafficking. {ECO:0000305}. | FUNCTION: Serine kinase that plays an essential role in the NF-kappa-B signaling pathway which is activated by multiple stimuli such as inflammatory cytokines, bacterial or viral products, DNA damages or other cellular stresses (PubMed:30337470). Acts as part of the canonical IKK complex in the conventional pathway of NF-kappa-B activation. Phosphorylates inhibitors of NF-kappa-B on 2 critical serine residues. These modifications allow polyubiquitination of the inhibitors and subsequent degradation by the proteasome. In turn, free NF-kappa-B is translocated into the nucleus and activates the transcription of hundreds of genes involved in immune response, growth control, or protection against apoptosis. In addition to the NF-kappa-B inhibitors, phosphorylates several other components of the signaling pathway including NEMO/IKBKG, NF-kappa-B subunits RELA and NFKB1, as well as IKK-related kinases TBK1 and IKBKE (PubMed:11297557, PubMed:20410276). IKK-related kinase phosphorylations may prevent the overproduction of inflammatory mediators since they exert a negative regulation on canonical IKKs. Phosphorylates FOXO3, mediating the TNF-dependent inactivation of this pro-apoptotic transcription factor (PubMed:15084260). Also phosphorylates other substrates including NCOA3, BCL10 and IRS1 (PubMed:17213322). Within the nucleus, acts as an adapter protein for NFKBIA degradation in UV-induced NF-kappa-B activation (PubMed:11297557). Phosphorylates RIPK1 at 'Ser-25' which represses its kinase activity and consequently prevents TNF-mediated RIPK1-dependent cell death (By similarity). Phosphorylates the C-terminus of IRF5, stimulating IRF5 homodimerization and translocation into the nucleus (PubMed:25326418). {ECO:0000250|UniProtKB:O88351, ECO:0000269|PubMed:11297557, ECO:0000269|PubMed:15084260, ECO:0000269|PubMed:17213322, ECO:0000269|PubMed:19716809, ECO:0000269|PubMed:20410276, ECO:0000269|PubMed:20434986, ECO:0000269|PubMed:20797629, ECO:0000269|PubMed:21138416, ECO:0000269|PubMed:25326418, ECO:0000269|PubMed:30337470}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HOOK3 | chr8:42798588 | chr8:42162705 | ENST00000307602 | + | 5 | 22 | 10_126 | 133.33333333333334 | 719.0 | Domain | Calponin-homology (CH) |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000416505 | 3 | 21 | 458_479 | 70.33333333333333 | 698.0 | Region | Note=Leucine-zipper | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000416505 | 3 | 21 | 737_742 | 70.33333333333333 | 698.0 | Region | Note=NEMO-binding | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000519735 | 4 | 9 | 458_479 | 129.33333333333334 | 257.0 | Region | Note=Leucine-zipper | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000519735 | 4 | 9 | 737_742 | 129.33333333333334 | 257.0 | Region | Note=NEMO-binding | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520810 | 4 | 22 | 458_479 | 129.33333333333334 | 757.0 | Region | Note=Leucine-zipper | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520810 | 4 | 22 | 737_742 | 129.33333333333334 | 757.0 | Region | Note=NEMO-binding | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520835 | 3 | 21 | 458_479 | 127.33333333333333 | 755.0 | Region | Note=Leucine-zipper | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520835 | 3 | 21 | 737_742 | 127.33333333333333 | 755.0 | Region | Note=NEMO-binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HOOK3 | chr8:42798588 | chr8:42162705 | ENST00000307602 | + | 5 | 22 | 167_433 | 133.33333333333334 | 719.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HOOK3 | chr8:42798588 | chr8:42162705 | ENST00000307602 | + | 5 | 22 | 462_667 | 133.33333333333334 | 719.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | HOOK3 | chr8:42798588 | chr8:42162705 | ENST00000307602 | + | 5 | 22 | 553_718 | 133.33333333333334 | 719.0 | Region | Note=Required for association with Golgi |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000416505 | 3 | 21 | 15_300 | 70.33333333333333 | 698.0 | Domain | Protein kinase | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000519735 | 4 | 9 | 15_300 | 129.33333333333334 | 257.0 | Domain | Protein kinase | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520810 | 4 | 22 | 15_300 | 129.33333333333334 | 757.0 | Domain | Protein kinase | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520835 | 3 | 21 | 15_300 | 127.33333333333333 | 755.0 | Domain | Protein kinase | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000416505 | 3 | 21 | 21_29 | 70.33333333333333 | 698.0 | Nucleotide binding | ATP | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000519735 | 4 | 9 | 21_29 | 129.33333333333334 | 257.0 | Nucleotide binding | ATP | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520810 | 4 | 22 | 21_29 | 129.33333333333334 | 757.0 | Nucleotide binding | ATP | |
| Tgene | IKBKB | chr8:42798588 | chr8:42162705 | ENST00000520835 | 3 | 21 | 21_29 | 127.33333333333333 | 755.0 | Nucleotide binding | ATP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| HOOK3 | CUL3, AKTIP, HOOK1, HOOK2, HOOK3, FAM160A2, VPS16, VPS41, SCFD1, CCDC8, ZC4H2, COMTD1, TNFSF13B, IFT57, FNTB, MRE11A, NTRK1, SSX2IP, CEP63, Nedd1, Rab5c, Zbtb48, DUSP3, FAM160A1, MTMR4, SYCE3, RALBP1, CDC16, NCAPH2, KLHL10, KLHL20, MYO6, TGOLN2, DYNC1I2, DYNC1LI1, DYNC1LI2, DYNC1H1, DCTN1, BRPF1, RAB21, RCN2, TPM3, MYH10, HYOU1, ACOT9, KRT19, ACTG1, ACACA, ACACB, IARS2, MTHFD1L, ZNF326, RAD50, MCCC1, GFPT1, PCCB, AASS, MTPAP, TUBB2B, HLCS, YWHAQ, MCM5, DLD, MYL6, ABCB7, PC, PCNA, CP, PCCA, PATL1, HNRNPH1, KRI1, SNX2, RBM14, DLG5, FAM160B1, FAM160B2, KIF1C, KLC4, TBC1D23, KIF1B, WDR11, FAM91A1, KLC2, GOLGA5, TBKBP1, WDR62, PCM1, NFKB1, TPD52L2, CPD, KIAA0319L, CEP192, PPP6R2, AAK1, KIF5B, ATG2A, CAPZA1, FAM21A, CAPZB, CCDC88A, FAM21C, NAP1L4, TJP2, ANKRD52, HAUS5, ABI2, GPATCH1, ANKRD28, KIAA0196, BSDC1, RELA, APC, FAM120A, LUZP1, COBL, BCR, BACH2, ARHGEF11, CEP55, WASF2, SIPA1L3, PPP6R3, AFTPH, VPS45, CORO1B, BCCIP, WDR83, PHF8, NAP1L1, LTV1, ALMS1, NBN, GAPVD1, OSBPL3, PPP6C, IFT27, CGN, SYNRG, MAP7D3, HBS1L, CEP97, ZC3H7A, DVL2, STRN, IGF2R, TJP1, SPAG5, CC2D1A, PLEC, MAPT, MEOX2, CTIF, KRT13, KRT27, PNMA5, ORF3a, M, nsp4, nsp6, ORF6, ORF7a, ORF7b, nsp12, nsp2, nsp3, CSDE1, TP53, EBAG9, AP4E1, KRT37, SYCE1, ITGA8, KIAA0226, HAP1, TMEM74, PIPSL, KRT31, FAM9C, NUBP1, CD40, MAGEA8, DUSP16, GPR17, KRT34, LCTL, AGBL4, SKAP1, ANKRD22, FEM1A, WHAMMP3, FTL, TRIM52, ARHGEF10, PPARD, SDF4, GFAP, EIF4ENIF1, Eif3g, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HOOK3 | 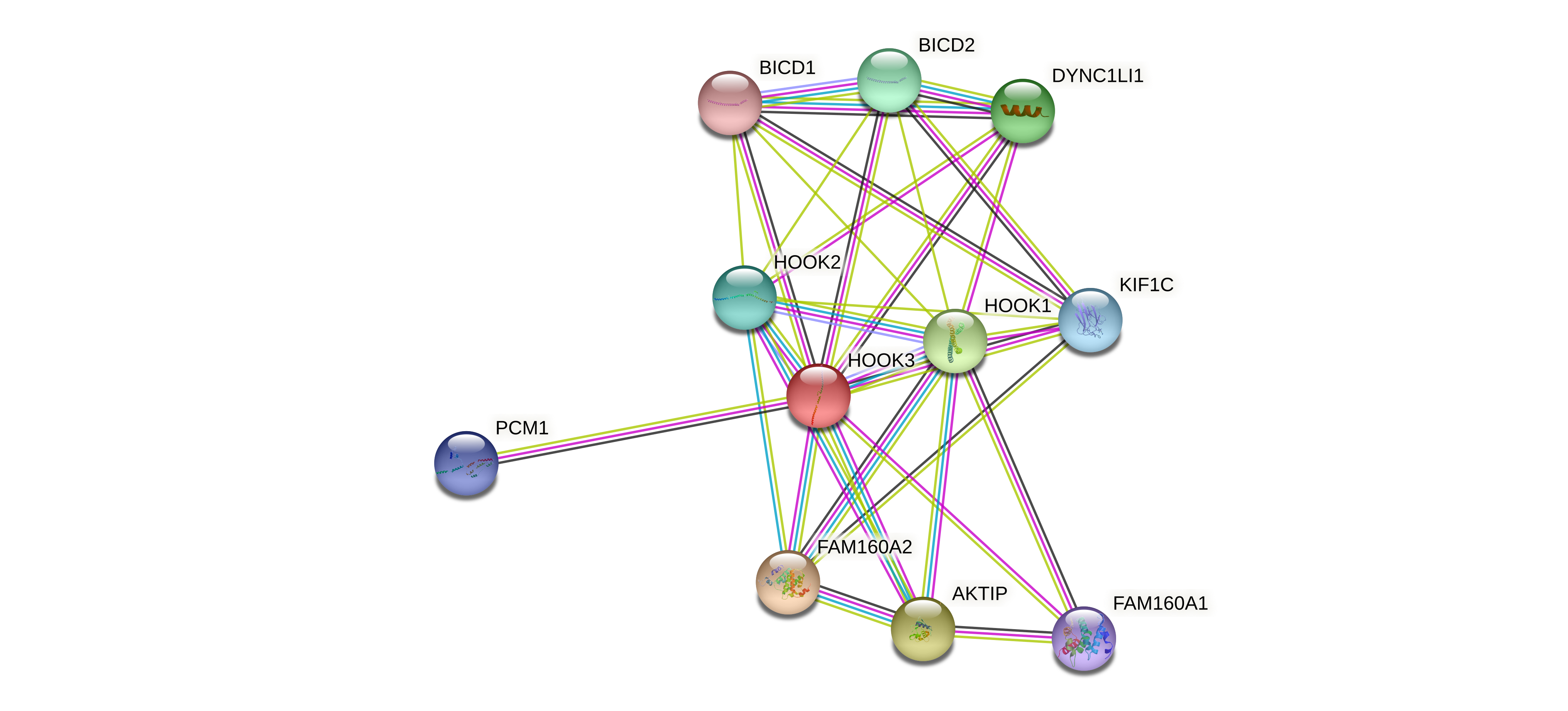 |
| IKBKB |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HOOK3-IKBKB |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HOOK3-IKBKB |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | HOOK3 | C0032460 | Polycystic Ovary Syndrome | 1 | CTD_human |
| Hgene | HOOK3 | C1136382 | Sclerocystic Ovaries | 1 | CTD_human |

