| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ITPR1-BHLHE40 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ITPR1-BHLHE40 | FusionPDB ID: 40677 | FusionGDB2.0 ID: 40677 | Hgene | Tgene | Gene symbol | ITPR1 | BHLHE40 | Gene ID | 3708 | 8553 |
| Gene name | inositol 1,4,5-trisphosphate receptor type 1 | basic helix-loop-helix family member e40 | |
| Synonyms | ACV|CLA4|INSP3R1|IP3R|IP3R1|PPP1R94|SCA15|SCA16|SCA29 | BHLHB2|Clast5|DEC1|HLHB2|SHARP-2|SHARP2|STRA13|Stra14 | |
| Cytomap | 3p26.1 | 3p26.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | inositol 1,4,5-trisphosphate receptor type 1IP3 receptorIP3R 1inositol 1,4,5-triphosphate receptor, type 1protein phosphatase 1, regulatory subunit 94type 1 InsP3 receptortype 1 inositol 1,4,5-trisphosphate receptor | class E basic helix-loop-helix protein 40basic helix-loop-helix domain containing, class B, 2class B basic helix-loop-helix protein 2differentially expressed in chondrocytes 1differentially expressed in chondrocytes protein 1differentiated embryo cho | |
| Modification date | 20200327 | 20200327 | |
| UniProtAcc | Q14643 | O14503 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000302640, ENST00000354582, ENST00000357086, ENST00000423119, ENST00000443694, ENST00000456211, ENST00000463980, ENST00000544951, | ENST00000256495, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 14 X 15 X 8=1680 | 3 X 4 X 3=36 |
| # samples | 16 | 4 | |
| ** MAII score | log2(16/1680*10)=-3.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ITPR1 [Title/Abstract] AND BHLHE40 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ITPR1(4704890)-BHLHE40(5023077), # samples:2 ITPR1(4808394)-BHLHE40(5024521), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ITPR1-BHLHE40 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ITPR1-BHLHE40 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ITPR1-BHLHE40 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ITPR1-BHLHE40 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ITPR1 | GO:0001666 | response to hypoxia | 19120137 |
| Hgene | ITPR1 | GO:0050849 | negative regulation of calcium-mediated signaling | 16793548 |
| Tgene | BHLHE40 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12397359|12624110|14672706|15193144 |
| Tgene | BHLHE40 | GO:0007623 | circadian rhythm | 28797635 |
| Tgene | BHLHE40 | GO:0032922 | circadian regulation of gene expression | 18411297 |
| Tgene | BHLHE40 | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 18411297 |
| Tgene | BHLHE40 | GO:0045892 | negative regulation of transcription, DNA-templated | 12397359|30012868 |
 Fusion gene breakpoints across ITPR1 (5'-gene) Fusion gene breakpoints across ITPR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
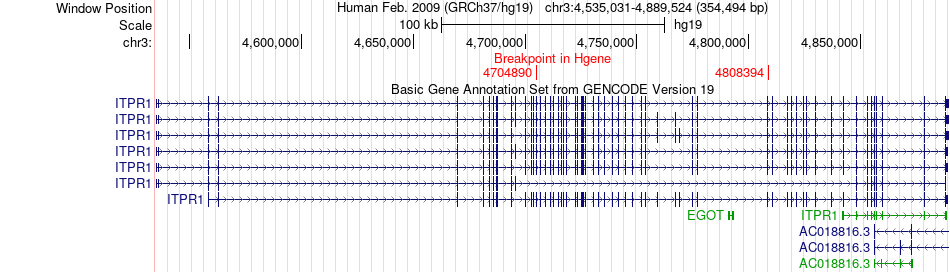 |
 Fusion gene breakpoints across BHLHE40 (3'-gene) Fusion gene breakpoints across BHLHE40 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
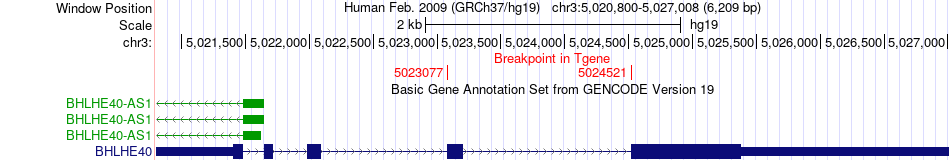 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DU-A7TA-01A | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + |
| ChimerDB4 | LGG | TCGA-DU-A7TA | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + |
| ChimerDB4 | UCEC | TCGA-D1-A3DH-01A | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000302640 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4471 | 1859 | 305 | 2839 | 844 |
| ENST00000354582 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4516 | 1904 | 305 | 2884 | 859 |
| ENST00000423119 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4516 | 1904 | 305 | 2884 | 859 |
| ENST00000456211 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4469 | 1857 | 303 | 2837 | 844 |
| ENST00000357086 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4514 | 1902 | 303 | 2882 | 859 |
| ENST00000443694 | ITPR1 | chr3 | 4704890 | + | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | 4121 | 1509 | 0 | 2489 | 829 |
| ENST00000302640 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8518 | 6030 | 305 | 6886 | 2193 |
| ENST00000354582 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8518 | 6030 | 305 | 6886 | 2193 |
| ENST00000423119 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8419 | 5931 | 305 | 6787 | 2160 |
| ENST00000456211 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8372 | 5884 | 303 | 6740 | 2145 |
| ENST00000357086 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8417 | 5929 | 303 | 6785 | 2160 |
| ENST00000443694 | ITPR1 | chr3 | 4808394 | + | ENST00000256495 | BHLHE40 | chr3 | 5024521 | + | 8168 | 5680 | 0 | 6536 | 2178 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000302640 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000562989 | 0.99943703 |
| ENST00000354582 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000422285 | 0.9995777 |
| ENST00000423119 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000422285 | 0.9995777 |
| ENST00000456211 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000550547 | 0.99944943 |
| ENST00000357086 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000412742 | 0.99958724 |
| ENST00000443694 | ENST00000256495 | ITPR1 | chr3 | 4704890 | + | BHLHE40 | chr3 | 5023077 | + | 0.000537878 | 0.9994622 |
| ENST00000302640 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.001413369 | 0.99858665 |
| ENST00000354582 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.001350467 | 0.99864954 |
| ENST00000423119 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.00154008 | 0.99845994 |
| ENST00000456211 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.001476718 | 0.9985233 |
| ENST00000357086 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.00152498 | 0.998475 |
| ENST00000443694 | ENST00000256495 | ITPR1 | chr3 | 4808394 | + | BHLHE40 | chr3 | 5024521 | + | 0.001050922 | 0.9989491 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >40677_40677_1_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000302640_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=844AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPE GNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDAS KVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEKAVVLELTLKHVKALT NLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKV MDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTK KNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMP -------------------------------------------------------------- >40677_40677_2_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000354582_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=859AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEK AVVLELTLKHVKALTNLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELL QGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGE RIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALS -------------------------------------------------------------- >40677_40677_3_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000357086_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=859AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEK AVVLELTLKHVKALTNLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELL QGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGE RIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALS -------------------------------------------------------------- >40677_40677_4_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000423119_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=859AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEK AVVLELTLKHVKALTNLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELL QGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGE RIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALS -------------------------------------------------------------- >40677_40677_5_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000443694_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=829AA_BP=0 MSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKAAKPGANSTTDAVLLN KLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQPFYKLRSIGDSVVIGD KVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDEHRKKQHVFLRTTGRQ SATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPEGNDISSIFELDPTTL RGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDASKVLGSIAGKLEKGTI TQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEKAVVLELTLKHVKALTNLIDQQQQKIIALQS GLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVMDFKEKPSSPAKGSE GPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKKNRMQLSDDEGHFTS SDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQRLPSPLPAHPSVDS -------------------------------------------------------------- >40677_40677_6_ITPR1-BHLHE40_ITPR1_chr3_4704890_ENST00000456211_BHLHE40_chr3_5023077_ENST00000256495_length(amino acids)=844AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPE GNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDAS KVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEKAVVLELTLKHVKALT NLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKV MDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTK KNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMP -------------------------------------------------------------- >40677_40677_7_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000302640_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2193AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPE GNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDAS KVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQAPFTDCGDGPMLRLEE LGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEIDTFVSLVRKNREPRF LDYLSDLCVSMNKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSNKEIRSKSVRELAQDA KEGQKEDRDVLSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDPQEQVTPVKYARLWSE IPSEIAIDDYDSSGASKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDLLRLTKILLAILDCVH VTTIFPISKMAKGEENKGNNDVEKLKSSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDIMVMDTKLKIIEILQF ILNVRLDYRISCLLCIFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGSEENTPLDLDDHGGRTFLRVLLHLTMH DYPPLVSGALQLLFRHFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDGASGENEHKKTEEGNN KPQKHESTSSYNYRVVKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMRLAHEFLQNFCAGNQQ NQALLHKHINLFLNPGILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAELVNSGED VLVFYNDRASFQTLIQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCIPEVKIAYINFLNHCY VDTEVEMKEIYTSNHMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQGVFRVYH CNWLMPSQKASVESCIRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRDYRNIIERLQDIVSAL EDRLRPLVQAELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKDRGYGEKLISIDELDN AELPPAPDSENATEELEPSPPLRQLEDHKRGEALRQVLVNRYYGNVRPSGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSL AEVQCHLDKEGASNLVIDLIMNASSDRVFHESILLAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDL GNKKKDDEVDRDAPSRKKGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVM DFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKK NRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQ -------------------------------------------------------------- >40677_40677_8_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000354582_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2193AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQA PFTDCGDGPMLRLEELGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEI DTFVSLVRKNREPRFLDYLSDLCVSMNKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSN KEIRSKSVRELAQDAKEGQKEDRDVLSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDP QEQVTPVKYARLWSEIPSEIAIDDYDSSGASKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDL LRLTKILLAILDCVHVTTIFPISKMAKGEENKGNNDVEKLKSSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDI MVMDTKLKIIEILQFILNVRLDYRISCLLCIFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGSEENTPLDLDDH GGRTFLRVLLHLTMHDYPPLVSGALQLLFRHFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDG ASGENEHKKTEEGNNKPQKHESTSSYNYRVVKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMR LAHEFLQNFCAGNQQNQALLHKHINLFLNPGILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKK CQDMVMAELVNSGEDVLVFYNDRASFQTLIQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCI PEVKIAYINFLNHCYVDTEVEMKEIYTSNHMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQ PVFVQLLQGVFRVYHCNWLMPSQKASVESCIRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRD YRNIIERLQDIVSALEDRLRPLVQAELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKD RGYGEKLISIDELDNAELPPAPDSENATEKGEALRQVLVNRYYGNVRPSGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSL AEVQCHLDKEGASNLVIDLIMNASSDRVFHESILLAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDL GNKKKDDEVDRDAPSRKKGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVM DFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKK NRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQ -------------------------------------------------------------- >40677_40677_9_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000357086_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2160AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQA PFTDCGDGPMLRLEELGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEI DTFVSLVRKNREPRFLDYLSDLCVSMNKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSN KEIRSKSVRELAQDAKEGQKEDRDVLSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDP QEQVTPVKYARLWSEIPSEIAIDDYDSSGASKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDL LRLTKILLAILDCVHVTTIFPISKMAKGEENKGSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDIMVMDTKLKI IEILQFILNVRLDYRISCLLCIFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGSEENTPLDLDDHGGRTFLRVL LHLTMHDYPPLVSGALQLLFRHFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDGASGENEHKK TEEGNNKPQKHESTSSYNYRVVKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMRLAHEFLQNF CAGNQQNQALLHKHINLFLNPGILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAEL VNSGEDVLVFYNDRASFQTLIQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCIPEVKIAYIN FLNHCYVDTEVEMKEIYTSNHMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQG VFRVYHCNWLMPSQKASVESCIRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRDYRNIIERLQ DIVSALEDRLRPLVQAELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKDRGYGEKGEA LRQVLVNRYYGNVRPSGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSLAEVQCHLDKEGASNLVIDLIMNASSDRVFHESI LLAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDLGNKKKDDEVDRDAPSRKKGELSGRNVETGQEMF CSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSG EQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFC LPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQRLPSPLPAHPSVDSSVLLQALKPIPPLNLETKD -------------------------------------------------------------- >40677_40677_10_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000423119_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2160AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQA PFTDCGDGPMLRLEELGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEI DTFVSLVRKNREPRFLDYLSDLCVSMNKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSN KEIRSKSVRELAQDAKEGQKEDRDVLSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDP QEQVTPVKYARLWSEIPSEIAIDDYDSSGASKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDL LRLTKILLAILDCVHVTTIFPISKMAKGEENKGSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDIMVMDTKLKI IEILQFILNVRLDYRISCLLCIFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGRKENTPLDLDDHGGRTFLRVL LHLTMHDYPPLVSGALQLLFRHFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDGASGENEHKK TEEGNNKPQKHESTSSYNYRVVKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMRLAHEFLQNF CAGNQQNQALLHKHINLFLNPGILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAEL VNSGEDVLVFYNDRASFQTLIQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCIPEVKIAYIN FLNHCYVDTEVEMKEIYTSNHMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQG VFRVYHCNWLMPSQKASVESCIRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRDYRNIIERLQ DIVSALEDRLRPLVQAELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKDRGYGEKGEA LRQVLVNRYYGNVRPSGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSLAEVQCHLDKEGASNLVIDLIMNASSDRVFHESI LLAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDLGNKKKDDEVDRDAPSRKKGELSGRNVETGQEMF CSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSG EQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFC LPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQRLPSPLPAHPSVDSSVLLQALKPIPPLNLETKD -------------------------------------------------------------- >40677_40677_11_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000443694_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2178AA_BP=0 MSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKAAKPGANSTTDAVLLN KLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQPFYKLRSIGDSVVIGD KVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDEHRKKQHVFLRTTGRQ SATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPEGNDISSIFELDPTTL RGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDASKVLGSIAGKLEKGTI TQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQAPFTDCGDGPMLRLEELGDQRHAPFRHICRL CYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEIDTFVSLVRKNREPRFLDYLSDLCVSMNKSI PVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSNKEIRSKSVRELAQDAKEGQKEDRDVLSYYR YQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDPQEQVTPVKYARLWSEIPSEIAIDDYDSSGA SKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDLLRLTKILLAILDCVHVTTIFPISKMAKGEE NKGNNDVEKLKSSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDIMVMDTKLKIIEILQFILNVRLDYRISCLLC IFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGSEENTPLDLDDHGGRTFLRVLLHLTMHDYPPLVSGALQLLFR HFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDGASGENEHKKTEEGNNKPQKHESTSSYNYRV VKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMRLAHEFLQNFCAGNQQNQALLHKHINLFLNP GILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAELVNSGEDVLVFYNDRASFQTLI QMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCIPEVKIAYINFLNHCYVDTEVEMKEIYTSNH MWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQGVFRVYHCNWLMPSQKASVESC IRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRDYRNIIERLQDIVSALEDRLRPLVQAELSVL VDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKDRGYGEKLISIDELDNAELPPAPDSENATEE LEPSPPLRQLEDHKRGEALRQVLVNRYYGNVRPSGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSLAEVQCHLDKEGASNL VIDLIMNASSDRVFHESILLAIALLEGGNTTIQHSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDLGNKKKDDEVDRDAPS RKKGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVMDFKEKPSSPAKGSEG PGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKKNRMQLSDDEGHFTSS DLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALSSFMNPDKISAPLLMPQRLPSPLPAHPSVDSS -------------------------------------------------------------- >40677_40677_12_ITPR1-BHLHE40_ITPR1_chr3_4808394_ENST00000456211_BHLHE40_chr3_5024521_ENST00000256495_length(amino acids)=2145AA_BP=1 MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDQDASRSRLRNAQEKMVYSLVSVPE GNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPVSPAEVRDLDFANDAS KVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQIFKLLQAPFTDCGDGPMLRLEE LGDQRHAPFRHICRLCYRVLRHSQQDYRKNQEYIAKQFGFMQKQIGYDVLAEDTITALLHNNRKLLEKHITAAEIDTFVSLVRKNREPRF LDYLSDLCVSMNKSIPVTQELICKAVLNPTNADILIETKLVLSRFEFEGVSSTGENALEAGEDEEEVWLFWRDSNKEIRSKSVRELAQDA KEGQKEDRDVLSYYRYQLNLFARMCLDRQYLAINEISGQLDVDLILRCMSDENLPYDLRASFCRLMLHMHVDRDPQEQVTPVKYARLWSE IPSEIAIDDYDSSGASKDEIKERFAQTMEFVEEYLRDVVCQRFPFSDKEKNKLTFEVVNLARNLIYFGFYNFSDLLRLTKILLAILDCVH VTTIFPISKMAKGEENKGSNVMRSIHGVGELMTQVVLRGGGFLPMTPMAAAPEGNVKQAEPEKEDIMVMDTKLKIIEILQFILNVRLDYR ISCLLCIFKREFDESNSQTSETSSGNSSQEGPSNVPGALDFEHIEEQAEGIFGGSEENTPLDLDDHGGRTFLRVLLHLTMHDYPPLVSGA LQLLFRHFSQRQEVLQAFKQVQLLVTSQDVDNYKQIKQDLDQLRSIVEKSELWVYKGQGPDETMDGASGENEHKKTEEGNNKPQKHESTS SYNYRVVKEILIRLSKLCVQESASVRKSRKQQQRLLRNMGAHAVVLELLQIPYEKAEDTKMQEIMRLAHEFLQNFCAGNQQNQALLHKHI NLFLNPGILEAVTMQHIFMNNFQLCSEINERVVQHFVHCIETHGRNVQYIKFLQTIVKAEGKFIKKCQDMVMAELVNSGEDVLVFYNDRA SFQTLIQMMRSERDRMDENSPLMYHIHLVELLAVCTEGKNVYTEIKCNSLLPLDDIVRVVTHEDCIPEVKIAYINFLNHCYVDTEVEMKE IYTSNHMWKLFENFLVDICRACNNTSDRKHADSILEKYVTEIVMSIVTTFFSSPFSDQSTTLQTRQPVFVQLLQGVFRVYHCNWLMPSQK ASVESCIRVLSDVAKSRAIAIPVDLDSQVNNLFLKSHSIVQKTAMNWRLSARNAARRDSVLAASRDYRNIIERLQDIVSALEDRLRPLVQ AELSVLVDVLHRPELLFPENTDARRKCESGGFICKLIKHTKQLLEENEEKLCIKVLQTLREMMTKDRGYGEKGEALRQVLVNRYYGNVRP SGRRESLTSFGNGPLSAGGPGKPGGGGGGSGSSSMSRGEMSLAEVQCHLDKEGASNLVIDLIMNASSDRVFHESILLAIALLEGGNTTIQ HSFFCRLTEDKKSEKFFKVFYDRMKVAQQEIKATVTVNTSDLGNKKKDDEVDRDAPSRKKGELSGRNVETGQEMFCSGFQTCAREVLQYL AKHENTRDLKSSQLVTHLHRVVSELLQGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGG ESEKGDLRSEQPCFKSDHGRRFTMGERIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:4704890/chr3:5023077) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ITPR1 | BHLHE40 |
| FUNCTION: Intracellular channel that mediates calcium release from the endoplasmic reticulum following stimulation by inositol 1,4,5-trisphosphate (PubMed:27108797). Involved in the regulation of epithelial secretion of electrolytes and fluid through the interaction with AHCYL1 (By similarity). Plays a role in ER stress-induced apoptosis. Cytoplasmic calcium released from the ER triggers apoptosis by the activation of CaM kinase II, eventually leading to the activation of downstream apoptosis pathways (By similarity). {ECO:0000250|UniProtKB:P11881, ECO:0000269|PubMed:27108797}. | FUNCTION: Transcriptional repressor involved in the regulation of the circadian rhythm by negatively regulating the activity of the clock genes and clock-controlled genes (PubMed:12397359, PubMed:18411297). Acts as the negative limb of a novel autoregulatory feedback loop (DEC loop) which differs from the one formed by the PER and CRY transcriptional repressors (PER/CRY loop) (PubMed:14672706). Both these loops are interlocked as it represses the expression of PER1/2 and in turn is repressed by PER1/2 and CRY1/2 (PubMed:15193144). Represses the activity of the circadian transcriptional activator: CLOCK-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer by competing for the binding to E-box elements (5'-CACGTG-3') found within the promoters of its target genes (PubMed:15560782). Negatively regulates its own expression and the expression of DBP and BHLHE41/DEC2 (PubMed:14672706). Acts as a corepressor of RXR and the RXR-LXR heterodimers and represses the ligand-induced RXRA and NR1H3/LXRA transactivation activity (PubMed:19786558). May be involved in the regulation of chondrocyte differentiation via the cAMP pathway (PubMed:19786558). Represses the transcription of NR0B2 and attentuates the transactivation of NR0B2 by the CLOCK-ARNTL/BMAL1 complex (PubMed:28797635). Drives the circadian rhythm of blood pressure through transcriptional repression of ATP1B1 in the cardiovascular system (PubMed:30012868). {ECO:0000269|PubMed:12397359, ECO:0000269|PubMed:14672706, ECO:0000269|PubMed:15193144, ECO:0000269|PubMed:15560782, ECO:0000269|PubMed:18411297, ECO:0000269|PubMed:19786558, ECO:0000269|PubMed:28797635, ECO:0000269|PubMed:30012868}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 112_166 | 503.0 | 2744.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 173_223 | 503.0 | 2744.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 231_287 | 503.0 | 2744.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 294_373 | 503.0 | 2744.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 379_435 | 503.0 | 2744.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 112_166 | 518.0 | 2711.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 173_223 | 518.0 | 2711.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 231_287 | 518.0 | 2711.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 294_373 | 518.0 | 2711.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 379_435 | 518.0 | 2711.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 112_166 | 503.0 | 2744.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 173_223 | 503.0 | 2744.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 231_287 | 503.0 | 2744.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 294_373 | 503.0 | 2744.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 379_435 | 503.0 | 2744.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 112_166 | 503.0 | 2696.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 173_223 | 503.0 | 2696.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 231_287 | 503.0 | 2696.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 294_373 | 503.0 | 2696.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 379_435 | 503.0 | 2696.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 112_166 | 1893.3333333333333 | 2744.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 173_223 | 1893.3333333333333 | 2744.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 231_287 | 1893.3333333333333 | 2744.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 294_373 | 1893.3333333333333 | 2744.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 379_435 | 1893.3333333333333 | 2744.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 112_166 | 1860.3333333333333 | 2711.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 173_223 | 1860.3333333333333 | 2711.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 231_287 | 1860.3333333333333 | 2711.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 294_373 | 1860.3333333333333 | 2711.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 379_435 | 1860.3333333333333 | 2711.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 112_166 | 1893.3333333333333 | 2744.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 173_223 | 1893.3333333333333 | 2744.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 231_287 | 1893.3333333333333 | 2744.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 294_373 | 1893.3333333333333 | 2744.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 379_435 | 1893.3333333333333 | 2744.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 112_166 | 1845.3333333333333 | 2696.0 | Domain | MIR 1 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 173_223 | 1845.3333333333333 | 2696.0 | Domain | MIR 2 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 231_287 | 1845.3333333333333 | 2696.0 | Domain | MIR 3 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 294_373 | 1845.3333333333333 | 2696.0 | Domain | MIR 4 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 379_435 | 1845.3333333333333 | 2696.0 | Domain | MIR 5 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 265_269 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 265_269 | 518.0 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 508_511 | 518.0 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 265_269 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 265_269 | 503.0 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 265_269 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 508_511 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 567_569 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 265_269 | 1860.3333333333333 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 508_511 | 1860.3333333333333 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 567_569 | 1860.3333333333333 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 265_269 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 508_511 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 567_569 | 1893.3333333333333 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 265_269 | 1845.3333333333333 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 508_511 | 1845.3333333333333 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 567_569 | 1845.3333333333333 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Tgene | BHLHE40 | chr3:4704890 | chr3:5023077 | ENST00000256495 | 2 | 5 | 142_175 | 86.0 | 413.0 | Domain | Orange | |
| Tgene | BHLHE40 | chr3:4808394 | chr3:5024521 | ENST00000256495 | 3 | 5 | 142_175 | 127.33333333333333 | 413.0 | Domain | Orange |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 508_511 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 567_569 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 567_569 | 518.0 | 2711.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 508_511 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 567_569 | 503.0 | 2744.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 508_511 | 503.0 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 567_569 | 503.0 | 2696.0 | Region | Inositol 1%2C4%2C5-trisphosphate binding |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 1_2282 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2304_2314 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2336_2361 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2383_2405 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2427_2448 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2470_2577 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2599_2758 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 1_2282 | 518.0 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2304_2314 | 518.0 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2336_2361 | 518.0 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2383_2405 | 518.0 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2427_2448 | 518.0 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2470_2577 | 518.0 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2599_2758 | 518.0 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 1_2282 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2304_2314 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2336_2361 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2383_2405 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2427_2448 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2470_2577 | 503.0 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2599_2758 | 503.0 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 1_2282 | 503.0 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2304_2314 | 503.0 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2336_2361 | 503.0 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2383_2405 | 503.0 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2427_2448 | 503.0 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2470_2577 | 503.0 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2599_2758 | 503.0 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 1_2282 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2304_2314 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2336_2361 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2383_2405 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2427_2448 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2470_2577 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2599_2758 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 1_2282 | 1860.3333333333333 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2304_2314 | 1860.3333333333333 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2336_2361 | 1860.3333333333333 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2383_2405 | 1860.3333333333333 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2427_2448 | 1860.3333333333333 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2470_2577 | 1860.3333333333333 | 2711.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2599_2758 | 1860.3333333333333 | 2711.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 1_2282 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2304_2314 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2336_2361 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2383_2405 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2427_2448 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2470_2577 | 1893.3333333333333 | 2744.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2599_2758 | 1893.3333333333333 | 2744.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 1_2282 | 1845.3333333333333 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2304_2314 | 1845.3333333333333 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2336_2361 | 1845.3333333333333 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2383_2405 | 1845.3333333333333 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2427_2448 | 1845.3333333333333 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2470_2577 | 1845.3333333333333 | 2696.0 | Topological domain | Lumenal |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2599_2758 | 1845.3333333333333 | 2696.0 | Topological domain | Cytoplasmic |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2283_2303 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2315_2335 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2362_2382 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2406_2426 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2449_2469 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2578_2598 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2283_2303 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2315_2335 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2362_2382 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2406_2426 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2449_2469 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2578_2598 | 518.0 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2283_2303 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2315_2335 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2362_2382 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2406_2426 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2449_2469 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2578_2598 | 503.0 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2283_2303 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2315_2335 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2362_2382 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2406_2426 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2449_2469 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2578_2598 | 503.0 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2283_2303 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2315_2335 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2362_2382 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2406_2426 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2449_2469 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2578_2598 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2283_2303 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2315_2335 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2362_2382 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2406_2426 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2449_2469 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2578_2598 | 1860.3333333333333 | 2711.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2283_2303 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2315_2335 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2362_2382 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2406_2426 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2449_2469 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2578_2598 | 1893.3333333333333 | 2744.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2283_2303 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2315_2335 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2362_2382 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2406_2426 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2449_2469 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2578_2598 | 1845.3333333333333 | 2696.0 | Transmembrane | Helical |
| Tgene | BHLHE40 | chr3:4704890 | chr3:5023077 | ENST00000256495 | 2 | 5 | 52_107 | 86.0 | 413.0 | Domain | bHLH | |
| Tgene | BHLHE40 | chr3:4808394 | chr3:5024521 | ENST00000256495 | 3 | 5 | 52_107 | 127.33333333333333 | 413.0 | Domain | bHLH |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1646_ITPR1_4704890_BHLHE40_5023077_ranked_0.pdb | ITPR1 | 4808394 | 4704890 | ENST00000256495 | BHLHE40 | chr3 | 5023077 | + | MATECLTQVRIFKKDMSDKMSSFLHIGDICSLYAEGSTNGFISTLGLVDDRCVVQPETGDLNNPPKKFRDCLFKLCPMNRYSAQKQFWKA AKPGANSTTDAVLLNKLHHAADLEKKQNETENRKLLGTVIQYGNVIQLLHLKSNKYLTVNKRLPALLEKNAMRVTLDEAGNEGSWFYIQP FYKLRSIGDSVVIGDKVVLNPVNAGQPLHASSHQLVDNPGCNEVNSVNCNTSWKIVLFMKWSDNKDDILKGGDVVRLFHAEQEKFLTCDE HRKKQHVFLRTTGRQSATSATSSKALWEVEVVQHDPCRGGAGYWNSLFRFKHLATGHYLAAEVDPDFEEECLEFQPSVDPDQDASRSRLR NAQEKMVYSLVSVPEGNDISSIFELDPTTLRGGDSLVPRNSYVRLRHLCTNTWVHSTNIPIDKEEEKPVMLKIGTSPVKEDKEAFAIVPV SPAEVRDLDFANDASKVLGSIAGKLEKGTITQNERRSVTKLLEDLVYFVTGGTNSGQDVLEVVFSKPNRERQKLMREQNILKQTLGHLEK AVVLELTLKHVKALTNLIDQQQQKIIALQSGLQAGELSGRNVETGQEMFCSGFQTCAREVLQYLAKHENTRDLKSSQLVTHLHRVVSELL QGGTSRKPSDPAPKVMDFKEKPSSPAKGSEGPGKNCVPVIQRTFAHSSGEQSGSDTDTDSGYGGESEKGDLRSEQPCFKSDHGRRFTMGE RIGAIKQESEEPPTKKNRMQLSDDEGHFTSSDLISSPFLGPHPHQPPFCLPFYLIPPSATAYLPMLEKCWYPTSVPVLYPGLNASAAALS | 859 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ITPR1_pLDDT.png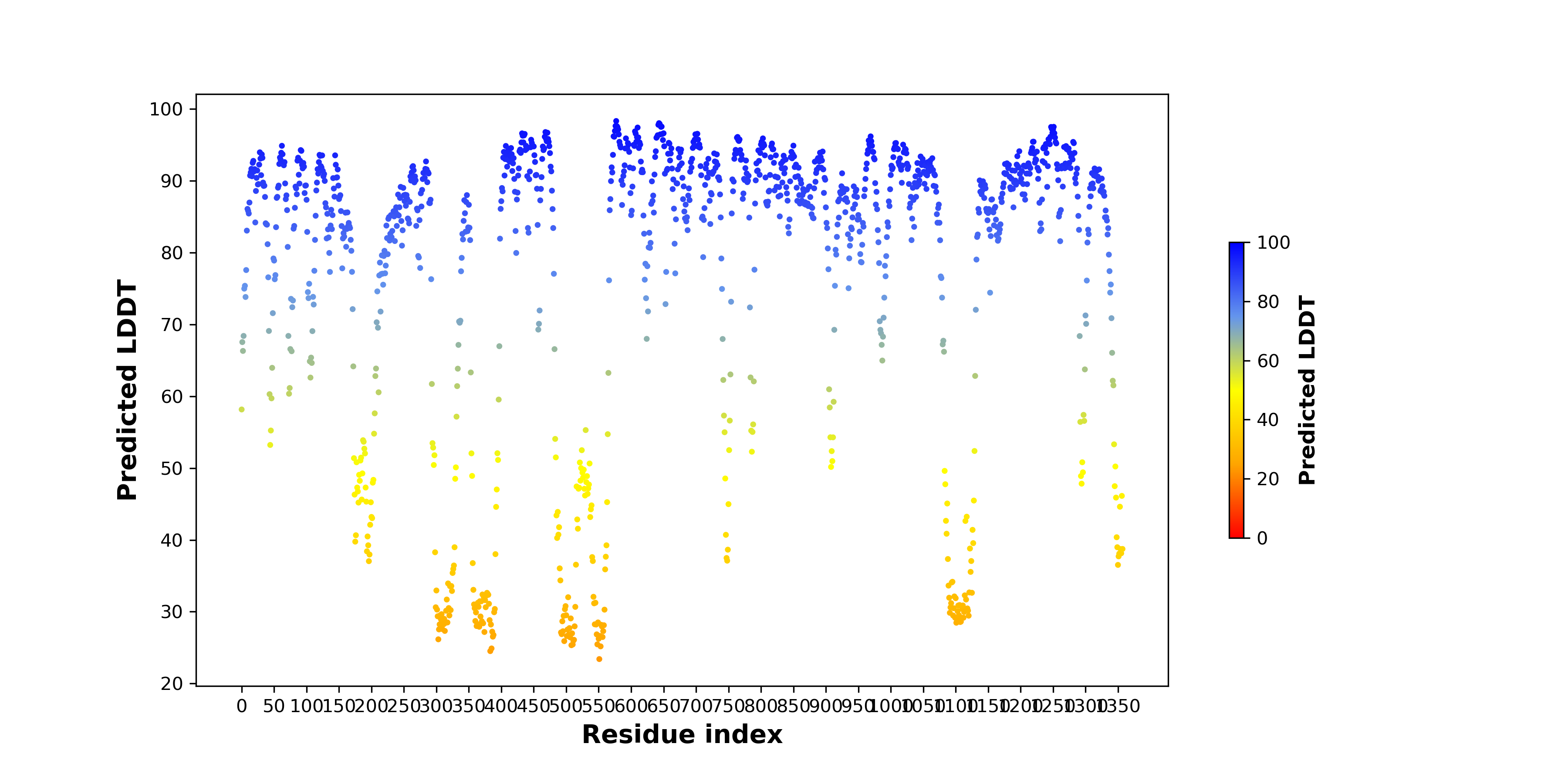 |
BHLHE40_pLDDT.png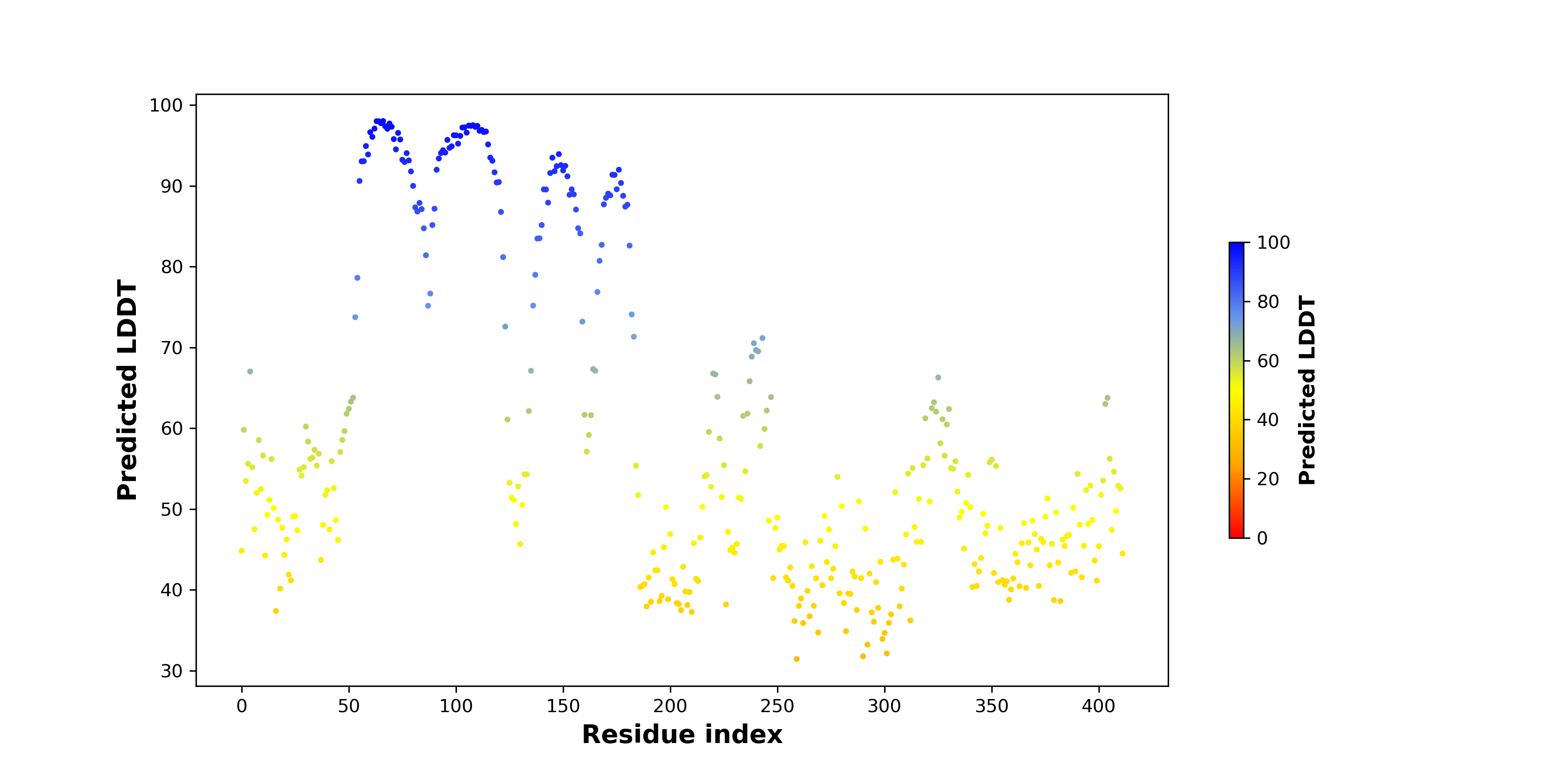 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
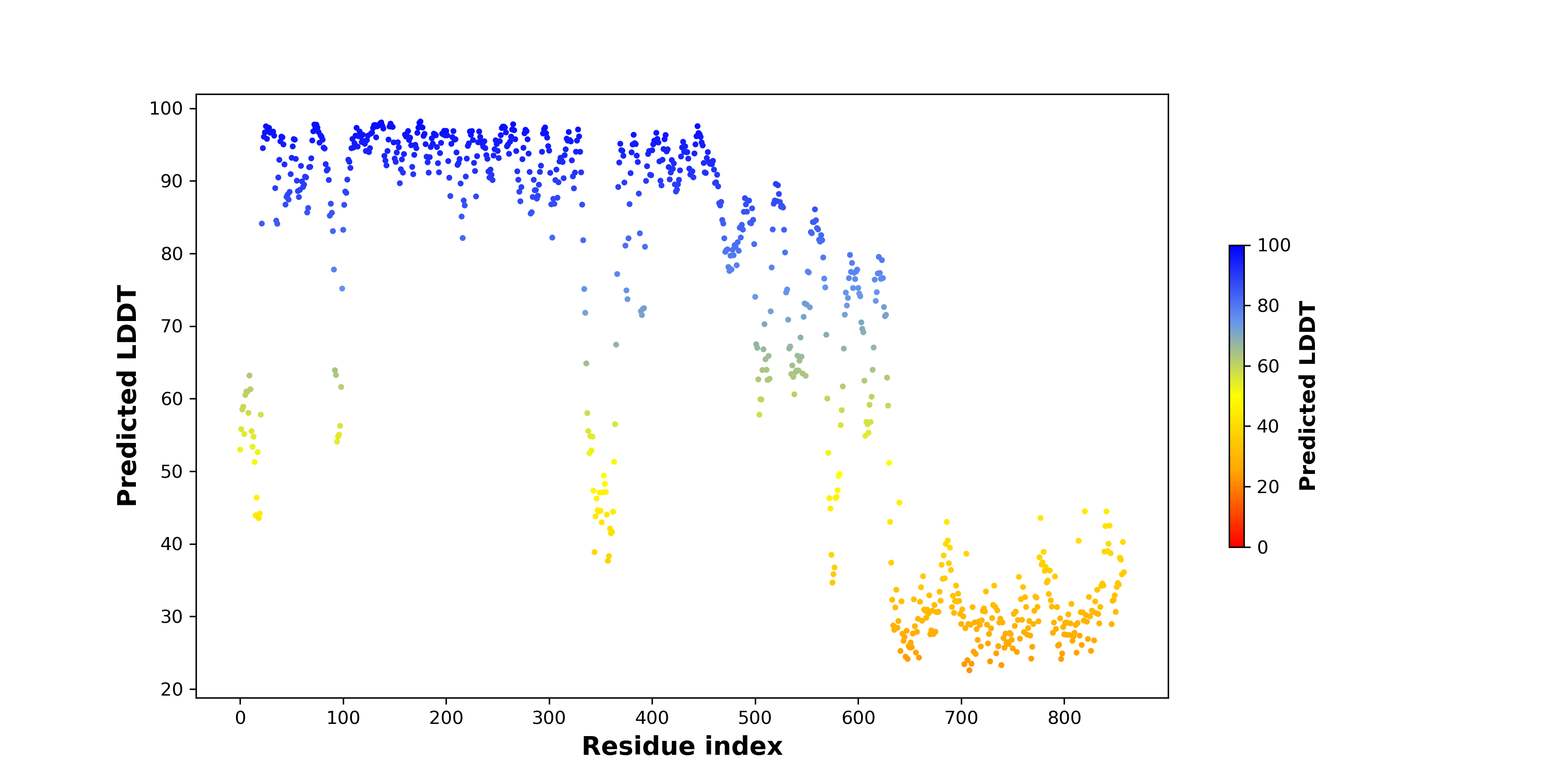 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ITPR1 | |
| BHLHE40 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000302640 | + | 15 | 61 | 2472_2537 | 503.0 | 2744.0 | ERP44 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000357086 | + | 16 | 59 | 2472_2537 | 518.0 | 2711.0 | ERP44 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000443694 | + | 13 | 59 | 2472_2537 | 503.0 | 2744.0 | ERP44 |
| Hgene | ITPR1 | chr3:4704890 | chr3:5023077 | ENST00000456211 | + | 15 | 58 | 2472_2537 | 503.0 | 2696.0 | ERP44 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000302640 | + | 44 | 61 | 2472_2537 | 1893.3333333333333 | 2744.0 | ERP44 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000357086 | + | 42 | 59 | 2472_2537 | 1860.3333333333333 | 2711.0 | ERP44 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000443694 | + | 42 | 59 | 2472_2537 | 1893.3333333333333 | 2744.0 | ERP44 |
| Hgene | ITPR1 | chr3:4808394 | chr3:5024521 | ENST00000456211 | + | 41 | 58 | 2472_2537 | 1845.3333333333333 | 2696.0 | ERP44 |
Top |
Related Drugs to ITPR1-BHLHE40 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ITPR1-BHLHE40 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

