| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:KCNC3-PRKDC |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: KCNC3-PRKDC | FusionPDB ID: 41380 | FusionGDB2.0 ID: 41380 | Hgene | Tgene | Gene symbol | KCNC3 | PRKDC | Gene ID | 3748 | 5591 |
| Gene name | potassium voltage-gated channel subfamily C member 3 | protein kinase, DNA-activated, catalytic subunit | |
| Synonyms | KSHIIID|KV3.3|SCA13 | DNA-PKC|DNA-PKcs|DNAPK|DNAPKc|DNPK1|HYRC|HYRC1|IMD26|XRCC7|p350 | |
| Cytomap | 19q13.33 | 8q11.21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | potassium voltage-gated channel subfamily C member 3Shaw-related voltage-gated potassium channel protein 3potassium channel, voltage gated Shaw related subfamily C, member 3potassium voltage-gated channel, Shaw-related subfamily, member 3voltage-gated | DNA-dependent protein kinase catalytic subunitDNA-PK catalytic subunithyper-radiosensitivity of murine scid mutation, complementing 1p460protein kinase, DNA-activated, catalytic polypeptide | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q14003 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000376959, ENST00000391818, ENST00000474951, ENST00000477616, | ENST00000523565, ENST00000314191, ENST00000338368, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 5 X 4 X 5=100 | 18 X 20 X 5=1800 |
| # samples | 5 | 19 | |
| ** MAII score | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1800*10)=-3.24392558288609 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: KCNC3 [Title/Abstract] AND PRKDC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KCNC3(50823850)-PRKDC(48730122), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | KCNC3-PRKDC seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNC3-PRKDC seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNC3-PRKDC seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KCNC3-PRKDC seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KCNC3-PRKDC seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KCNC3-PRKDC seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. KCNC3-PRKDC seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KCNC3-PRKDC seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNC3 | GO:0051262 | protein tetramerization | 23734863 |
| Tgene | PRKDC | GO:0002218 | activation of innate immune response | 28712728 |
| Tgene | PRKDC | GO:0006468 | protein phosphorylation | 26237645 |
| Tgene | PRKDC | GO:0006974 | cellular response to DNA damage stimulus | 26237645 |
| Tgene | PRKDC | GO:0018105 | peptidyl-serine phosphorylation | 15194694|19303849 |
| Tgene | PRKDC | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 26237645 |
 Fusion gene breakpoints across KCNC3 (5'-gene) Fusion gene breakpoints across KCNC3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
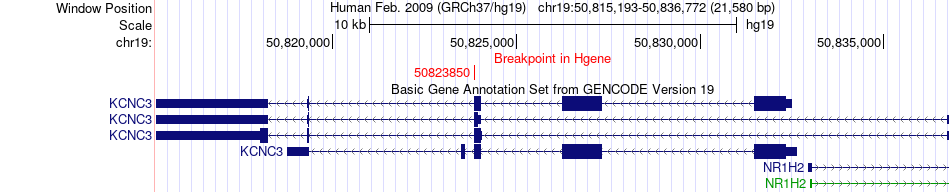 |
 Fusion gene breakpoints across PRKDC (3'-gene) Fusion gene breakpoints across PRKDC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
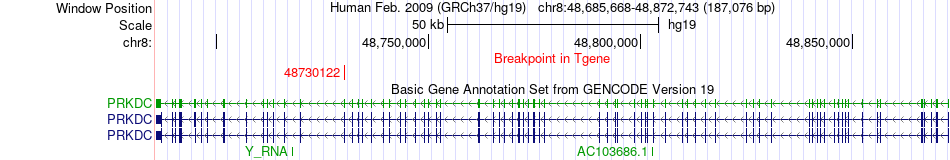 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | CESC | TCGA-VS-A8EI-01A | KCNC3 | chr19 | 50823850 | - | PRKDC | chr8 | 48730122 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000376959 | KCNC3 | chr19 | 50823850 | - | ENST00000338368 | PRKDC | chr8 | 48730122 | - | 6248 | 2334 | 128 | 5182 | 1684 |
| ENST00000376959 | KCNC3 | chr19 | 50823850 | - | ENST00000314191 | PRKDC | chr8 | 48730122 | - | 6341 | 2334 | 128 | 5275 | 1715 |
| ENST00000477616 | KCNC3 | chr19 | 50823850 | - | ENST00000338368 | PRKDC | chr8 | 48730122 | - | 6379 | 2465 | 259 | 5313 | 1684 |
| ENST00000477616 | KCNC3 | chr19 | 50823850 | - | ENST00000314191 | PRKDC | chr8 | 48730122 | - | 6472 | 2465 | 259 | 5406 | 1715 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000376959 | ENST00000338368 | KCNC3 | chr19 | 50823850 | - | PRKDC | chr8 | 48730122 | - | 0.001336683 | 0.9986633 |
| ENST00000376959 | ENST00000314191 | KCNC3 | chr19 | 50823850 | - | PRKDC | chr8 | 48730122 | - | 0.001050846 | 0.9989492 |
| ENST00000477616 | ENST00000338368 | KCNC3 | chr19 | 50823850 | - | PRKDC | chr8 | 48730122 | - | 0.001516597 | 0.9984835 |
| ENST00000477616 | ENST00000314191 | KCNC3 | chr19 | 50823850 | - | PRKDC | chr8 | 48730122 | - | 0.001197198 | 0.9988028 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >41380_41380_1_KCNC3-PRKDC_KCNC3_chr19_50823850_ENST00000376959_PRKDC_chr8_48730122_ENST00000314191_length(amino acids)=1715AA_BP=735 MRPRPSRPAPRPMLSSVCVSSFRGRQGASKQQPAPPPQPPESPPPPPLPPQQQQPAQPGPAASPAGPPAPRGPGDRRAEPCPGLPAAAMG RHGGGGGDSGKIVINVGGVRHETYRSTLRTLPGTRLAGLTEPEAAARFDYDPGADEFFFDRHPGVFAYVLNYYRTGKLHCPADVCGPLFE EELGFWGIDETDVEACCWMTYRQHRDAEEALDSFEAPDPAGAANAANAAGAHDGGLDDEAGAGGGGLDGAGGELKRLCFQDAGGGAGGPP GGAGGAGGTWWRRWQPRVWALFEDPYSSRAARYVAFASLFFILISITTFCLETHEGFIHISNKTVTQASPIPGAPPENITNVEVETEPFL TYVEGVCVVWFTFEFLMRITFCPDKVEFLKSSLNIIDCVAILPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTRHFVGLRVLG HTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGADPDDILGSNHTYFKNIPIGFWWAVVTMTTLGYGDMYPKTWSGMLVGALCALAGV LTIAMPVPVIVNNFGMYYSLAMAKQKLPKKKNKHIPRPPQPGSPNYCKPDPPPPPPPHPHHGSGGISPPPPITPPSMGVTVAGAYPAGPH THPGLLRGGAGGLGIMGLPPLPAPGEPCPLAQEEVIEINRADPRPNGDPAAAALAHEDCPAIDQPAMSPEDKSPITPGSRGRYSRDRACF LLTDYAPSPDGSIRKGNLSSQVPLKRLLNTWTNRYPDAKMDPMNIWDDIITNRCFFLSKIEEKLTPLPEDNSMNVDQDGDPSDRMEVQEQ EEDISSLIRSCKFSMKMKMIDSARKQNNFSLAMKLLKELHKESKTRDDWLVSWVQSYCRLSHCRSRSQGCSEQVLTVLKTVSLLDENNVS SYLSKNILAFRDQNILLGTTYRIIANALSSEPACLAEIEEDKARRILELSGSSSEDSEKVIAGLYQRAFQHLSEAVQAAEEEAQPPSWSC GPAAGVIDAYMTLADFCDQQLRKEEENASVIDSAELQAYPALVVEKMLKALKLNSNEARLKFPRLLQIIERYPEETLSLMTKEISSVPCW QFISWISHMVALLDKDQAVAVQHSVEEITDNYPQAIVYPFIISSESYSFKDTSTGHKNKEFVARIKSKLDQGGVIQDFINALDQLSNPEL LFKDWSNDVRAELAKTPVNKKNIEKMYERMYAALGDPKAPGLGAFRRKFIQTFGKEFDKHFGKGGSKLLRMKLSDFNDITNMLLLKMNKD SKPPGNLKECSPWMSDFKVEFLRNELEIPGQYDGRGKPLPEYHVRIAGFDERVTVMASLRRPKRIIIRGHDEREHPFLVKGGEDLRQDQR VEQLFQVMNGILAQDSACSQRALQLRTYSVVPMTSRLGLIEWLENTVTLKDLLLNTMSQEEKAAYLSDPRAPPCEYKDWLTKMSGKHDVG AYMLMYKGANRTETVTSFRKRESKVPADLLKRAFVRMSTSPEAFLALRSHFASSHALICISHWILGIGDRHLNNFMVAMETGGVIGIDFG HAFGSATQFLPVPELMPFRLTRQFINLMLPMKETGLMYSIMVHALRAFRSDPGLLTNTMDVFVKEPSFDWKNFEQKMLKKGGSWIQEINV AEKNWYPRQKICYAKRKLAGANPAVITCDELLLGHEKAPAFRDYVAVARGSKDHNIRAQEPESGLSEETQVKCLMDQATDPNILGRTWEG -------------------------------------------------------------- >41380_41380_2_KCNC3-PRKDC_KCNC3_chr19_50823850_ENST00000376959_PRKDC_chr8_48730122_ENST00000338368_length(amino acids)=1684AA_BP=735 MRPRPSRPAPRPMLSSVCVSSFRGRQGASKQQPAPPPQPPESPPPPPLPPQQQQPAQPGPAASPAGPPAPRGPGDRRAEPCPGLPAAAMG RHGGGGGDSGKIVINVGGVRHETYRSTLRTLPGTRLAGLTEPEAAARFDYDPGADEFFFDRHPGVFAYVLNYYRTGKLHCPADVCGPLFE EELGFWGIDETDVEACCWMTYRQHRDAEEALDSFEAPDPAGAANAANAAGAHDGGLDDEAGAGGGGLDGAGGELKRLCFQDAGGGAGGPP GGAGGAGGTWWRRWQPRVWALFEDPYSSRAARYVAFASLFFILISITTFCLETHEGFIHISNKTVTQASPIPGAPPENITNVEVETEPFL TYVEGVCVVWFTFEFLMRITFCPDKVEFLKSSLNIIDCVAILPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTRHFVGLRVLG HTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGADPDDILGSNHTYFKNIPIGFWWAVVTMTTLGYGDMYPKTWSGMLVGALCALAGV LTIAMPVPVIVNNFGMYYSLAMAKQKLPKKKNKHIPRPPQPGSPNYCKPDPPPPPPPHPHHGSGGISPPPPITPPSMGVTVAGAYPAGPH THPGLLRGGAGGLGIMGLPPLPAPGEPCPLAQEEVIEINRADPRPNGDPAAAALAHEDCPAIDQPAMSPEDKSPITPGSRGRYSRDRACF LLTDYAPSPDGSIRKGNLSSQVPLKRLLNTWTNRYPDAKMDPMNIWDDIITNRCFFLSKIEEKLTPLPEDNSMNVDQDGDPSDRMEVQEQ EEDISSLIRSCKFSMKMKMIDSARKQNNFSLAMKLLKELHKESKTRDDWLVSWVQSYCRLSHCRSRSQGCSEQVLTVLKTVSLLDENNVS SYLSKNILAFRDQNILLGTTYRIIANALSSEPACLAEIEEDKARRILELSGSSSEDSEKVIAGLYQRAFQHLSEAVQAAEEEAQPPSWSC GPAAGVIDAYMTLADFCDQQLRKEEENASVIDSAELQAYPALVVEKMLKALKLNSNEARLKFPRLLQIIERYPEETLSLMTKEISSVPCW QFISWISHMVALLDKDQAVAVQHSVEEITDNYPQAIVYPFIISSESYSFKDTSTGHKNKEFVARIKSKLDQGGVIQDFINALDQLSNPEL LFKDWSNDVRAELAKTPVNKKNIEKMYERMYAALGDPKAPGLGAFRRKFIQTFGKEFDKHFGKGGSKLLRMKLSDFNDITNMLLLKMNKD SKPPGNLKECSPWMSDFKVEFLRNELEIPGQYDGRGKPLPEYHVRIAGFDERVTVMASLRRPKRIIIRGHDEREHPFLVKGGEDLRQDQR VEQLFQVMNGILAQDSACSQRALQLRTYSVVPMTSSDPRAPPCEYKDWLTKMSGKHDVGAYMLMYKGANRTETVTSFRKRESKVPADLLK RAFVRMSTSPEAFLALRSHFASSHALICISHWILGIGDRHLNNFMVAMETGGVIGIDFGHAFGSATQFLPVPELMPFRLTRQFINLMLPM KETGLMYSIMVHALRAFRSDPGLLTNTMDVFVKEPSFDWKNFEQKMLKKGGSWIQEINVAEKNWYPRQKICYAKRKLAGANPAVITCDEL -------------------------------------------------------------- >41380_41380_3_KCNC3-PRKDC_KCNC3_chr19_50823850_ENST00000477616_PRKDC_chr8_48730122_ENST00000314191_length(amino acids)=1715AA_BP=735 MRPRPSRPAPRPMLSSVCVSSFRGRQGASKQQPAPPPQPPESPPPPPLPPQQQQPAQPGPAASPAGPPAPRGPGDRRAEPCPGLPAAAMG RHGGGGGDSGKIVINVGGVRHETYRSTLRTLPGTRLAGLTEPEAAARFDYDPGADEFFFDRHPGVFAYVLNYYRTGKLHCPADVCGPLFE EELGFWGIDETDVEACCWMTYRQHRDAEEALDSFEAPDPAGAANAANAAGAHDGGLDDEAGAGGGGLDGAGGELKRLCFQDAGGGAGGPP GGAGGAGGTWWRRWQPRVWALFEDPYSSRAARYVAFASLFFILISITTFCLETHEGFIHISNKTVTQASPIPGAPPENITNVEVETEPFL TYVEGVCVVWFTFEFLMRITFCPDKVEFLKSSLNIIDCVAILPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTRHFVGLRVLG HTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGADPDDILGSNHTYFKNIPIGFWWAVVTMTTLGYGDMYPKTWSGMLVGALCALAGV LTIAMPVPVIVNNFGMYYSLAMAKQKLPKKKNKHIPRPPQPGSPNYCKPDPPPPPPPHPHHGSGGISPPPPITPPSMGVTVAGAYPAGPH THPGLLRGGAGGLGIMGLPPLPAPGEPCPLAQEEVIEINRADPRPNGDPAAAALAHEDCPAIDQPAMSPEDKSPITPGSRGRYSRDRACF LLTDYAPSPDGSIRKGNLSSQVPLKRLLNTWTNRYPDAKMDPMNIWDDIITNRCFFLSKIEEKLTPLPEDNSMNVDQDGDPSDRMEVQEQ EEDISSLIRSCKFSMKMKMIDSARKQNNFSLAMKLLKELHKESKTRDDWLVSWVQSYCRLSHCRSRSQGCSEQVLTVLKTVSLLDENNVS SYLSKNILAFRDQNILLGTTYRIIANALSSEPACLAEIEEDKARRILELSGSSSEDSEKVIAGLYQRAFQHLSEAVQAAEEEAQPPSWSC GPAAGVIDAYMTLADFCDQQLRKEEENASVIDSAELQAYPALVVEKMLKALKLNSNEARLKFPRLLQIIERYPEETLSLMTKEISSVPCW QFISWISHMVALLDKDQAVAVQHSVEEITDNYPQAIVYPFIISSESYSFKDTSTGHKNKEFVARIKSKLDQGGVIQDFINALDQLSNPEL LFKDWSNDVRAELAKTPVNKKNIEKMYERMYAALGDPKAPGLGAFRRKFIQTFGKEFDKHFGKGGSKLLRMKLSDFNDITNMLLLKMNKD SKPPGNLKECSPWMSDFKVEFLRNELEIPGQYDGRGKPLPEYHVRIAGFDERVTVMASLRRPKRIIIRGHDEREHPFLVKGGEDLRQDQR VEQLFQVMNGILAQDSACSQRALQLRTYSVVPMTSRLGLIEWLENTVTLKDLLLNTMSQEEKAAYLSDPRAPPCEYKDWLTKMSGKHDVG AYMLMYKGANRTETVTSFRKRESKVPADLLKRAFVRMSTSPEAFLALRSHFASSHALICISHWILGIGDRHLNNFMVAMETGGVIGIDFG HAFGSATQFLPVPELMPFRLTRQFINLMLPMKETGLMYSIMVHALRAFRSDPGLLTNTMDVFVKEPSFDWKNFEQKMLKKGGSWIQEINV AEKNWYPRQKICYAKRKLAGANPAVITCDELLLGHEKAPAFRDYVAVARGSKDHNIRAQEPESGLSEETQVKCLMDQATDPNILGRTWEG -------------------------------------------------------------- >41380_41380_4_KCNC3-PRKDC_KCNC3_chr19_50823850_ENST00000477616_PRKDC_chr8_48730122_ENST00000338368_length(amino acids)=1684AA_BP=735 MRPRPSRPAPRPMLSSVCVSSFRGRQGASKQQPAPPPQPPESPPPPPLPPQQQQPAQPGPAASPAGPPAPRGPGDRRAEPCPGLPAAAMG RHGGGGGDSGKIVINVGGVRHETYRSTLRTLPGTRLAGLTEPEAAARFDYDPGADEFFFDRHPGVFAYVLNYYRTGKLHCPADVCGPLFE EELGFWGIDETDVEACCWMTYRQHRDAEEALDSFEAPDPAGAANAANAAGAHDGGLDDEAGAGGGGLDGAGGELKRLCFQDAGGGAGGPP GGAGGAGGTWWRRWQPRVWALFEDPYSSRAARYVAFASLFFILISITTFCLETHEGFIHISNKTVTQASPIPGAPPENITNVEVETEPFL TYVEGVCVVWFTFEFLMRITFCPDKVEFLKSSLNIIDCVAILPFYLEVGLSGLSSKAAKDVLGFLRVVRFVRILRIFKLTRHFVGLRVLG HTLRASTNEFLLLIIFLALGVLIFATMIYYAERIGADPDDILGSNHTYFKNIPIGFWWAVVTMTTLGYGDMYPKTWSGMLVGALCALAGV LTIAMPVPVIVNNFGMYYSLAMAKQKLPKKKNKHIPRPPQPGSPNYCKPDPPPPPPPHPHHGSGGISPPPPITPPSMGVTVAGAYPAGPH THPGLLRGGAGGLGIMGLPPLPAPGEPCPLAQEEVIEINRADPRPNGDPAAAALAHEDCPAIDQPAMSPEDKSPITPGSRGRYSRDRACF LLTDYAPSPDGSIRKGNLSSQVPLKRLLNTWTNRYPDAKMDPMNIWDDIITNRCFFLSKIEEKLTPLPEDNSMNVDQDGDPSDRMEVQEQ EEDISSLIRSCKFSMKMKMIDSARKQNNFSLAMKLLKELHKESKTRDDWLVSWVQSYCRLSHCRSRSQGCSEQVLTVLKTVSLLDENNVS SYLSKNILAFRDQNILLGTTYRIIANALSSEPACLAEIEEDKARRILELSGSSSEDSEKVIAGLYQRAFQHLSEAVQAAEEEAQPPSWSC GPAAGVIDAYMTLADFCDQQLRKEEENASVIDSAELQAYPALVVEKMLKALKLNSNEARLKFPRLLQIIERYPEETLSLMTKEISSVPCW QFISWISHMVALLDKDQAVAVQHSVEEITDNYPQAIVYPFIISSESYSFKDTSTGHKNKEFVARIKSKLDQGGVIQDFINALDQLSNPEL LFKDWSNDVRAELAKTPVNKKNIEKMYERMYAALGDPKAPGLGAFRRKFIQTFGKEFDKHFGKGGSKLLRMKLSDFNDITNMLLLKMNKD SKPPGNLKECSPWMSDFKVEFLRNELEIPGQYDGRGKPLPEYHVRIAGFDERVTVMASLRRPKRIIIRGHDEREHPFLVKGGEDLRQDQR VEQLFQVMNGILAQDSACSQRALQLRTYSVVPMTSSDPRAPPCEYKDWLTKMSGKHDVGAYMLMYKGANRTETVTSFRKRESKVPADLLK RAFVRMSTSPEAFLALRSHFASSHALICISHWILGIGDRHLNNFMVAMETGGVIGIDFGHAFGSATQFLPVPELMPFRLTRQFINLMLPM KETGLMYSIMVHALRAFRSDPGLLTNTMDVFVKEPSFDWKNFEQKMLKKGGSWIQEINVAEKNWYPRQKICYAKRKLAGANPAVITCDEL -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:50823850/chr8:48730122) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KCNC3 | . |
| FUNCTION: Voltage-gated potassium channel that plays an important role in the rapid repolarization of fast-firing brain neurons. The channel opens in response to the voltage difference across the membrane, forming a potassium-selective channel through which potassium ions pass in accordance with their electrochemical gradient. The channel displays rapid activation and inactivation kinetics (PubMed:10712820, PubMed:26997484, PubMed:22289912, PubMed:23734863, PubMed:16501573, PubMed:19953606, PubMed:21479265, PubMed:25756792). It plays a role in the regulation of the frequency, shape and duration of action potentials in Purkinje cells. Required for normal survival of cerebellar neurons, probably via its role in regulating the duration and frequency of action potentials that in turn regulate the activity of voltage-gated Ca(2+) channels and cellular Ca(2+) homeostasis (By similarity). Required for normal motor function (PubMed:23734863, PubMed:16501573, PubMed:19953606, PubMed:21479265, PubMed:25756792). Plays a role in the reorganization of the cortical actin cytoskeleton and the formation of actin veil structures in neuronal growth cones via its interaction with HAX1 and the Arp2/3 complex (PubMed:26997484). {ECO:0000250|UniProtKB:Q63959, ECO:0000269|PubMed:10712820, ECO:0000269|PubMed:16501573, ECO:0000269|PubMed:19953606, ECO:0000269|PubMed:21479265, ECO:0000269|PubMed:22289912, ECO:0000269|PubMed:23734863, ECO:0000269|PubMed:25756792, ECO:0000269|PubMed:26997484}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 229_234 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 31_38 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Pro |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 39_42 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Gln |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 577_587 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Pro |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 596_599 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Pro |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 668_673 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Ala |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 81_85 | 723.3333333333334 | 237.33333333333334 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 503_508 | 723.3333333333334 | 237.33333333333334 | Motif | Selectivity filter |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 1_290 | 723.3333333333334 | 237.33333333333334 | Topological domain | Cytoplasmic |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 371_379 | 723.3333333333334 | 237.33333333333334 | Topological domain | Cytoplasmic |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 435_447 | 723.3333333333334 | 237.33333333333334 | Topological domain | Cytoplasmic |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 540_757 | 723.3333333333334 | 237.33333333333334 | Topological domain | Cytoplasmic |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 291_309 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 351_370 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 380_398 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 412_434 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Voltage-sensor%3B Name%3DSegment S4 |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 448_469 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNC3 | chr19:50823850 | chr8:48730122 | ENST00000477616 | - | 3 | 5 | 518_539 | 723.3333333333334 | 237.33333333333334 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 3747_4015 | 3147.3333333333335 | 4128.0 | Domain | PI3K/PI4K | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 4096_4128 | 3147.3333333333335 | 4128.0 | Domain | FATC |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 2906_3539 | 3147.3333333333335 | 4128.0 | Domain | FAT | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 1503_1538 | 3147.3333333333335 | 4128.0 | Region | Note=Leucine-zipper | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 2436_3212 | 3147.3333333333335 | 4128.0 | Region | Note=KIP-binding | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 1004_1040 | 3147.3333333333335 | 4128.0 | Repeat | Note=HEAT 2 | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 1723_1756 | 3147.3333333333335 | 4128.0 | Repeat | Note=TPR 1 | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 288_323 | 3147.3333333333335 | 4128.0 | Repeat | Note=HEAT 1 | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 2920_2948 | 3147.3333333333335 | 4128.0 | Repeat | Note=TPR 2 | |
| Tgene | PRKDC | chr19:50823850 | chr8:48730122 | ENST00000314191 | 67 | 87 | 2949_2982 | 3147.3333333333335 | 4128.0 | Repeat | Note=TPR 3 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| KCNC3 | |
| PRKDC |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to KCNC3-PRKDC |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to KCNC3-PRKDC |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

