| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:KCNMA1-DOCK1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: KCNMA1-DOCK1 | FusionPDB ID: 41484 | FusionGDB2.0 ID: 41484 | Hgene | Tgene | Gene symbol | KCNMA1 | DOCK1 | Gene ID | 3778 | 1793 |
| Gene name | potassium calcium-activated channel subfamily M alpha 1 | dedicator of cytokinesis 1 | |
| Synonyms | BKTM|CADEDS|IEG16|KCa1.1|LIWAS|MaxiK|PNKD3|SAKCA|SLO|SLO-ALPHA|SLO1|bA205K10.1|hSlo|mSLO1 | DOCK180|ced5 | |
| Cytomap | 10q22.3 | 10q26.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium-activated potassium channel subunit alpha-1uncharacterized proteinBK channel alpha subunitBKCA alpha subunitbig potassium channel alpha subunitcalcium-activated potassium channel, subfamily M subunit alpha-1k(VCA)alphamaxi-K channel HSLOpo | dedicator of cytokinesis protein 1180 kDa protein downstream of CRKDOwnstream of CrK | |
| Modification date | 20200315 | 20200327 | |
| UniProtAcc | Q12791 | Q5JSL3 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000286627, ENST00000286628, ENST00000354353, ENST00000372440, ENST00000372443, ENST00000404771, ENST00000404857, ENST00000406533, ENST00000480683, ENST00000481070, ENST00000484507, | ENST00000484400, ENST00000280333, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 20 X 17 X 10=3400 | 16 X 15 X 8=1920 |
| # samples | 25 | 16 | |
| ** MAII score | log2(25/3400*10)=-3.76553474636298 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1920*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: KCNMA1 [Title/Abstract] AND DOCK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KCNMA1(78880731)-DOCK1(129249596), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | KCNMA1-DOCK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNMA1-DOCK1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KCNMA1-DOCK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KCNMA1-DOCK1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | KCNMA1 | GO:0001666 | response to hypoxia | 15528406 |
| Hgene | KCNMA1 | GO:0006813 | potassium ion transport | 7573516|7877450|11245614|12388065|17706472|18458941 |
| Hgene | KCNMA1 | GO:0006970 | response to osmotic stress | 10840032|12388065 |
| Hgene | KCNMA1 | GO:0030007 | cellular potassium ion homeostasis | 11245614 |
| Hgene | KCNMA1 | GO:0034465 | response to carbon monoxide | 15528406 |
| Hgene | KCNMA1 | GO:0042391 | regulation of membrane potential | 7877450|7993625 |
| Hgene | KCNMA1 | GO:0045794 | negative regulation of cell volume | 12388065 |
| Hgene | KCNMA1 | GO:0051592 | response to calcium ion | 12388065|18458941 |
| Hgene | KCNMA1 | GO:0060073 | micturition | 11641143 |
| Hgene | KCNMA1 | GO:0060083 | smooth muscle contraction involved in micturition | 11641143 |
 Fusion gene breakpoints across KCNMA1 (5'-gene) Fusion gene breakpoints across KCNMA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
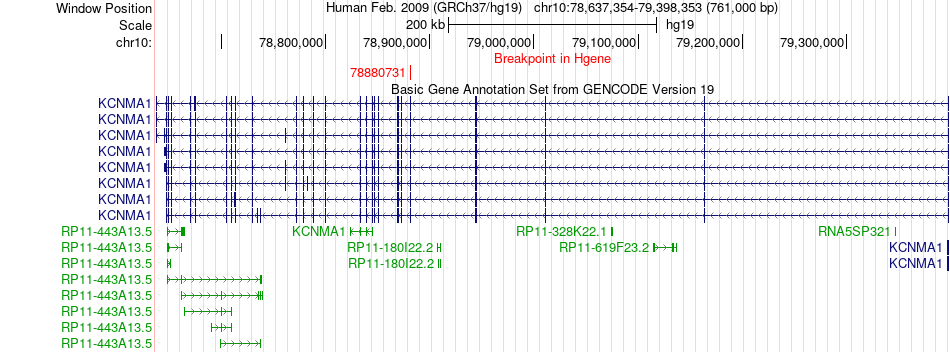 |
 Fusion gene breakpoints across DOCK1 (3'-gene) Fusion gene breakpoints across DOCK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
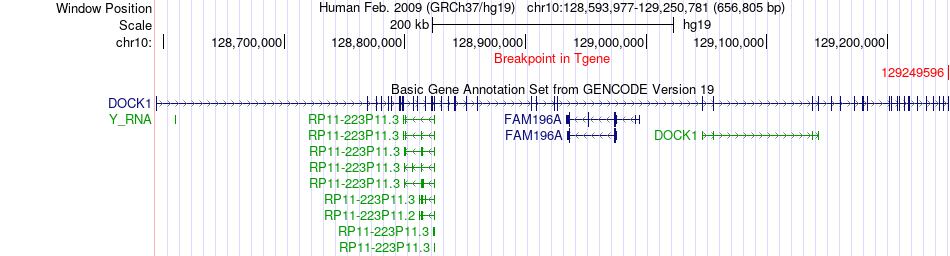 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-CC-A7IJ-01A | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000404771 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2186 | 1000 | 62 | 1267 | 401 |
| ENST00000372440 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2421 | 1235 | 297 | 1502 | 401 |
| ENST00000372443 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2797 | 1611 | 673 | 1878 | 401 |
| ENST00000286628 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000286627 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 3023 | 1837 | 899 | 2104 | 401 |
| ENST00000354353 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000404857 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
| ENST00000406533 | KCNMA1 | chr10 | 78880731 | - | ENST00000280333 | DOCK1 | chr10 | 129249596 | + | 2070 | 884 | 0 | 1151 | 383 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000404771 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.035057057 | 0.96494293 |
| ENST00000372440 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.044519797 | 0.9554802 |
| ENST00000372443 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.04783887 | 0.95216113 |
| ENST00000286628 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000286627 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.055738155 | 0.94426185 |
| ENST00000354353 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000404857 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
| ENST00000406533 | ENST00000280333 | KCNMA1 | chr10 | 78880731 | - | DOCK1 | chr10 | 129249596 | + | 0.03677205 | 0.963228 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >41484_41484_1_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286627_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_2_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000286628_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_3_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000354353_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_4_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372440_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_5_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000372443_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_6_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404771_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=401AA_BP=312 MAAGGAPPSPCAGCPLLAMANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIP VTMEVPCDSRGQRMWWAFLASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSV KDWAGVMISAQTLTGRVLVVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDF FTVPPVFVSVYLNRSWLGLRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPIS -------------------------------------------------------------- >41484_41484_7_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000404857_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- >41484_41484_8_KCNMA1-DOCK1_KCNMA1_chr10_78880731_ENST00000406533_DOCK1_chr10_129249596_ENST00000280333_length(amino acids)=383AA_BP=294 MANGGGGGGGSSGGGGGGGGSSLRMSSNIHANHLSLDASSSSSSSSSSSSSSSSSSSSSSVHEPKMDALIIPVTMEVPCDSRGQRMWWAF LASSMVTFFGGLFIILLWRTLKYLWTVCCHCGGKTKEAQKINNGSSQADGTLKPVDEKEEAVAAEVGWMTSVKDWAGVMISAQTLTGRVL VVLVFALSIGALVIYFIDSSNPIESCQNFYKDFTLQIDMAFNVFFLLYFGLRFIAANDKLWFWLEVNSVVDFFTVPPVFVSVYLNRSWLG LRFLRALRLIQFSEILQFLNILKTSICHLHCPAKLRLLPLQRQLASRHRWTPGSCSDVASLSGKSVLPLPISMPSPSVSPESAVYLIGPV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:78880731/chr10:129249596) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KCNMA1 | DOCK1 |
| FUNCTION: Potassium channel activated by both membrane depolarization or increase in cytosolic Ca(2+) that mediates export of K(+) (PubMed:29330545, PubMed:31152168). It is also activated by the concentration of cytosolic Mg(2+). Its activation dampens the excitatory events that elevate the cytosolic Ca(2+) concentration and/or depolarize the cell membrane. It therefore contributes to repolarization of the membrane potential. Plays a key role in controlling excitability in a number of systems, such as regulation of the contraction of smooth muscle, the tuning of hair cells in the cochlea, regulation of transmitter release, and innate immunity. In smooth muscles, its activation by high level of Ca(2+), caused by ryanodine receptors in the sarcoplasmic reticulum, regulates the membrane potential. In cochlea cells, its number and kinetic properties partly determine the characteristic frequency of each hair cell and thereby helps to establish a tonotopic map. Kinetics of KCNMA1 channels are determined by alternative splicing, phosphorylation status and its combination with modulating beta subunits. Highly sensitive to both iberiotoxin (IbTx) and charybdotoxin (CTX). {ECO:0000269|PubMed:29330545, ECO:0000269|PubMed:31152168}. | FUNCTION: Guanine nucleotide-exchange factor (GEF) that activates CDC42 by exchanging bound GDP for free GTP. Required for marginal zone (MZ) B-cell development, is associated with early bone marrow B-cell development, MZ B-cell formation, MZ B-cell number and marginal metallophilic macrophages morphology. Facilitates filopodia formation through the activation of CDC42. {ECO:0000250|UniProtKB:A2AF47}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 13_20 | 294.6666666666667 | 1179.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 39_60 | 294.6666666666667 | 1179.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 4_10 | 294.6666666666667 | 1179.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 13_20 | 294.6666666666667 | 1237.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 39_60 | 294.6666666666667 | 1237.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 4_10 | 294.6666666666667 | 1237.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 13_20 | 294.6666666666667 | 1220.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 39_60 | 294.6666666666667 | 1220.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 4_10 | 294.6666666666667 | 1220.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 108_178 | 294.6666666666667 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1_86 | 294.6666666666667 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 200_214 | 294.6666666666667 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 236_239 | 294.6666666666667 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 261_264 | 294.6666666666667 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 108_178 | 294.6666666666667 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1_86 | 294.6666666666667 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 200_214 | 294.6666666666667 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 236_239 | 294.6666666666667 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 261_264 | 294.6666666666667 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 108_178 | 294.6666666666667 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1_86 | 294.6666666666667 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 200_214 | 294.6666666666667 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 236_239 | 294.6666666666667 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 261_264 | 294.6666666666667 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 179_199 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 215_235 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 240_260 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 265_285 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 87_107 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 179_199 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 215_235 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 240_260 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 265_285 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 87_107 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 179_199 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 215_235 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 240_260 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 265_285 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 87_107 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 13_20 | 0 | 169.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 39_60 | 0 | 169.0 | Compositional bias | Note=Poly-Ser |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 4_10 | 0 | 169.0 | Compositional bias | Note=Poly-Gly |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 415_558 | 294.6666666666667 | 1179.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 415_558 | 294.6666666666667 | 1237.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 415_558 | 294.6666666666667 | 1220.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 415_558 | 0 | 169.0 | Domain | Note=RCK N-terminal |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 336_358 | 294.6666666666667 | 1179.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 336_358 | 294.6666666666667 | 1237.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 336_358 | 294.6666666666667 | 1220.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 336_358 | 0 | 169.0 | Intramembrane | Pore-forming%3B Name%3DP region |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1003_1025 | 294.6666666666667 | 1179.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 352_355 | 294.6666666666667 | 1179.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1003_1025 | 294.6666666666667 | 1237.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 352_355 | 294.6666666666667 | 1237.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1003_1025 | 294.6666666666667 | 1220.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 352_355 | 294.6666666666667 | 1220.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1003_1025 | 0 | 169.0 | Motif | Note=Calcium bowl |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 352_355 | 0 | 169.0 | Motif | Note=Selectivity for potassium |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 1032_1052 | 294.6666666666667 | 1179.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 556_576 | 294.6666666666667 | 1179.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 613_633 | 294.6666666666667 | 1179.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 677_681 | 294.6666666666667 | 1179.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 837_857 | 294.6666666666667 | 1179.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 1032_1052 | 294.6666666666667 | 1237.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 556_576 | 294.6666666666667 | 1237.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 613_633 | 294.6666666666667 | 1237.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 677_681 | 294.6666666666667 | 1237.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 837_857 | 294.6666666666667 | 1237.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 1032_1052 | 294.6666666666667 | 1220.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 556_576 | 294.6666666666667 | 1220.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 613_633 | 294.6666666666667 | 1220.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 677_681 | 294.6666666666667 | 1220.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 837_857 | 294.6666666666667 | 1220.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1032_1052 | 0 | 169.0 | Region | Note=Segment S10 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 556_576 | 0 | 169.0 | Region | Note=Segment S7 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 613_633 | 0 | 169.0 | Region | Note=Segment S8 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 677_681 | 0 | 169.0 | Region | Note=Heme-binding motif |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 837_857 | 0 | 169.0 | Region | Note=Segment S9 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 286_300 | 294.6666666666667 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 322_335 | 294.6666666666667 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 359_367 | 294.6666666666667 | 1179.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 389_1236 | 294.6666666666667 | 1179.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 286_300 | 294.6666666666667 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 322_335 | 294.6666666666667 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 359_367 | 294.6666666666667 | 1237.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 389_1236 | 294.6666666666667 | 1237.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 286_300 | 294.6666666666667 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 322_335 | 294.6666666666667 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 359_367 | 294.6666666666667 | 1220.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 389_1236 | 294.6666666666667 | 1220.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 108_178 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 1_86 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 200_214 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 236_239 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 261_264 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 286_300 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 322_335 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 359_367 | 0 | 169.0 | Topological domain | Extracellular |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 389_1236 | 0 | 169.0 | Topological domain | Cytoplasmic |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 301_321 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286627 | - | 6 | 27 | 368_388 | 294.6666666666667 | 1179.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 301_321 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000286628 | - | 6 | 28 | 368_388 | 294.6666666666667 | 1237.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 301_321 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000404857 | - | 6 | 28 | 368_388 | 294.6666666666667 | 1220.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 179_199 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S1 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 215_235 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S2 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 240_260 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S3 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 265_285 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S4 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 301_321 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S5 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 368_388 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S6 |
| Hgene | KCNMA1 | chr10:78880731 | chr10:129249596 | ENST00000480683 | - | 1 | 2 | 87_107 | 0 | 169.0 | Transmembrane | Helical%3B Name%3DSegment S0 |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1207_1617 | 1834.0 | 1866.0 | Domain | DOCKER | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 425_609 | 1834.0 | 1866.0 | Domain | C2 DOCK-type | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 9_70 | 1834.0 | 1866.0 | Domain | SH3 | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1687_1695 | 1834.0 | 1866.0 | Region | Phosphoinositide-binding |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| KCNMA1 | |
| DOCK1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1793_1819 | 1834.0 | 1866.0 | NCK2 second and third SH3 domain (minor) | |
| Tgene | DOCK1 | chr10:78880731 | chr10:129249596 | ENST00000280333 | 50 | 52 | 1820_1836 | 1834.0 | 1866.0 | NCK2 third SH3 domain (major) |
Top |
Related Drugs to KCNMA1-DOCK1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to KCNMA1-DOCK1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

