| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:LAMA3-OSBPL1A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: LAMA3-OSBPL1A | FusionPDB ID: 43877 | FusionGDB2.0 ID: 43877 | Hgene | Tgene | Gene symbol | LAMA3 | OSBPL1A | Gene ID | 3910 | 114876 |
| Gene name | laminin subunit alpha 4 | oxysterol binding protein like 1A | |
| Synonyms | CMD1JJ|LAMA3|LAMA4*-1 | ORP-1|ORP1|OSBPL1B | |
| Cytomap | 6q21 | 18q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | laminin subunit alpha-4laminin alpha 4 chainlaminin, alpha 4 | oxysterol-binding protein-related protein 1OSBP-related protein 1oxysterol binding protein-like 1Boxysterol-binding protein-related protein 1 variant 1oxysterol-binding protein-related protein 1 variant 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q16787 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000588770, ENST00000269217, ENST00000313654, ENST00000399516, ENST00000587184, | ENST00000319481, ENST00000357041, ENST00000399441, ENST00000399443, ENST00000582618, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 28 X 24 X 10=6720 | 18 X 15 X 10=2700 |
| # samples | 33 | 19 | |
| ** MAII score | log2(33/6720*10)=-4.34792330342031 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/2700*10)=-3.82888808360725 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: LAMA3 [Title/Abstract] AND OSBPL1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | LAMA3(21513899)-OSBPL1A(21892070), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | LAMA3-OSBPL1A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LAMA3-OSBPL1A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LAMA3-OSBPL1A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LAMA3-OSBPL1A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LAMA3 (5'-gene) Fusion gene breakpoints across LAMA3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
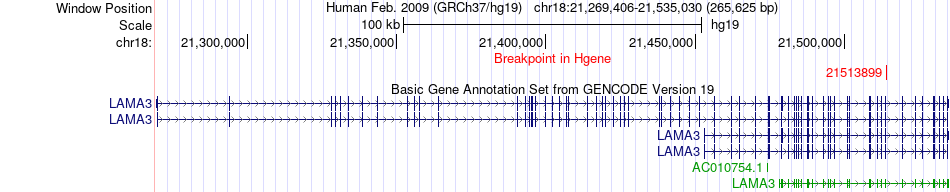 |
 Fusion gene breakpoints across OSBPL1A (3'-gene) Fusion gene breakpoints across OSBPL1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
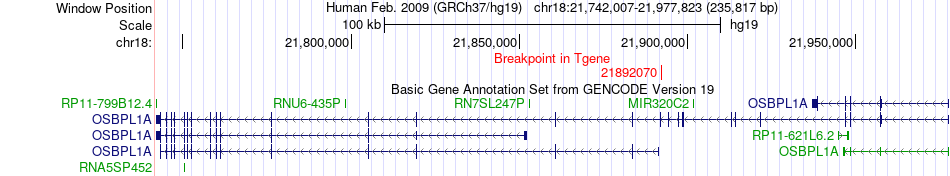 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-IN-A7NU-01A | LAMA3 | chr18 | 21513899 | - | OSBPL1A | chr18 | 21892070 | - |
| ChimerDB4 | STAD | TCGA-IN-A7NU-01A | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - |
| ChimerDB4 | STAD | TCGA-IN-A7NU | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000313654 | LAMA3 | chr18 | 21513899 | + | ENST00000319481 | OSBPL1A | chr18 | 21892070 | - | 12122 | 9103 | 241 | 10986 | 3581 |
| ENST00000399516 | LAMA3 | chr18 | 21513899 | + | ENST00000319481 | OSBPL1A | chr18 | 21892070 | - | 11713 | 8694 | 0 | 10577 | 3525 |
| ENST00000269217 | LAMA3 | chr18 | 21513899 | + | ENST00000319481 | OSBPL1A | chr18 | 21892070 | - | 7259 | 4240 | 205 | 6123 | 1972 |
| ENST00000587184 | LAMA3 | chr18 | 21513899 | + | ENST00000319481 | OSBPL1A | chr18 | 21892070 | - | 6886 | 3867 | 0 | 5750 | 1916 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000313654 | ENST00000319481 | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - | 0.000473404 | 0.9995266 |
| ENST00000399516 | ENST00000319481 | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - | 0.00033432 | 0.99966574 |
| ENST00000269217 | ENST00000319481 | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - | 0.000636219 | 0.99936384 |
| ENST00000587184 | ENST00000319481 | LAMA3 | chr18 | 21513899 | + | OSBPL1A | chr18 | 21892070 | - | 0.000492782 | 0.9995072 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >43877_43877_1_LAMA3-OSBPL1A_LAMA3_chr18_21513899_ENST00000269217_OSBPL1A_chr18_21892070_ENST00000319481_length(amino acids)=1972AA_BP=1344 MPPAVRRSACSMGWLWIFGAALGQCLGYSSQQQRVPFLQPPGQSQLQASYVEFRPSQGCSPGYYRDHKGLYTGRCVPCNCNGHSNQCQDG SGICVNCQHNTAGEHCERCQEGYYGNAVHGSCRACPCPHTNSFATGCVVNGGDVRCSCKAGYTGTQCERCAPGYFGNPQKFGGSCQPCSC NSNGQLGSCHPLTGDCINQEPKDSSPAEECDDCDSCVMTLLNDLATMGEQLRLVKSQLQGLSASAGLLEQMRHMETQAKDLRNQLLNYRS AISNHGSKIEGLERELTDLNQEFETLQEKAQVNSRKAQTLNNNVNRATQSAKELDVKIKNVIRNVHILLKQISGTDGEGNNVPSGDFSRE WAEAQRMMRELRNRNFGKHLREAEADKRESQLLLNRIRTWQKTHQGENNGLANSIRDSLNEYEAKLSDLRARLQEAAAQAKQANGLNQEN ERALGAIQRQVKEINSLQSDFTKYLTTADSSLLQTNIALQLMEKSQKEYEKLAASLNEARQELSDKVRELSRSAGKTSLVEEAEKHARSL QELAKQLEEIKRNASGDELVRCAVDAATAYENILNAIKAAEDAANRAASASESALQTVIKEDLPRKAKTLSSNSDKLLNEAKMTQKKLKQ EVSPALNNLQQTLNIVTVQKEVIDTNLTTLRDGLHGIQRGDIDAMISSAKSMVRKANDITDEVLDGLNPIQTDVERIKDTYGRTQNEDFK KALTDADNSVNKLTNKLPDLWRKIESINQQLLPLGNISDNMDRIRELIQQARDAASKVAVPMRFNGKSGVEVRLPNDLEDLKGYTSLSLF LQRPNSRENGGTENMFVMYLGNKDASRDYIGMAVVDGQLTCVYNLGDREAELQVDQILTKSETKEAVMDRVKFQRIYQFARLNYTKGATS SKPETPGVYDMDGRNSNTLLNLDPENVVFYVGGYPPDFKLPSRLSFPPYKGCIELDDLNENVLSLYNFKKTFNLNTTEVEPCRRRKEESD KNYFEGTGYARVPTQPHAPIPTFGQTIQTTVDRGLLFFAENGDRFISLNIEDGKLMVRYKLNSELPKERGVGDAINNGRDHSIQIKIGKL QKRMWINVDVQNTIIDGEVFDFSTYYLGGIPIAIRERFNISTPAFRGCMKNLKKTSGVVRLNDTVGVTKKCSEDWKLVRSASFSRGGQLS FTDLGLPPTDHLQASFGFQTFQPSGILLDHQTWTRNLQVTLEDGYIELSTSDSGSPIFKSPQTYMDGLLHYVSVISDNSGLRLLIDDQLL RNSKRLKHISSSRQSLRLGGSNFEGCISNVFVQRLSLSPEVLDLTSNSLKRDVSLGGCSLNKPPFLMLLKGSTRFNKTKTFRINQDWLEA IEEHSAYSTHYCSQDQLTDEEEEDTVSAADLKKSLEKAQSCQQRLDREISNFLKMIKECDMAKEMLPSFLQKVEVVSEASRETCVALTDC LNLFTKQEGVRNFKLEQEQEKNKILSEALETLATEHHELEQSLVKGSPPASILSEDEFYDALSDSESERSLSRLEAVTARSFEEEGEHLG SRKHRMSEEKDCGGGDALSNGIKKHRTSLPSPMFSRNDFSIWSILRKCIGMELSKITMPVIFNEPLSFLQRLTEYMEHTYLIHKASSLSD PVERMQCVAAFAVSAVASQWERTGKPFNPLLGETYELVRDDLGFRLISEQVSHHPPISAFHAEGLNNDFIFHGSIYPKLKFWGKSVEAEP KGTITLELLEHNEAYTWTNPTCCVHNIIVGKLWIEQYGNVEIINHKTGDKCVLNFKPCGLFGKELHKVEGYIQDKSKKKLCALYGKWTEC LYSVDPATFDAYKKNDKKNTEEKKNSKQMSTSEELDEMPVPDSESVFIIPGSVLLWRIAPRPPNSAQMYNFTSFAMVLNEVDKDMESVIP -------------------------------------------------------------- >43877_43877_2_LAMA3-OSBPL1A_LAMA3_chr18_21513899_ENST00000313654_OSBPL1A_chr18_21892070_ENST00000319481_length(amino acids)=3581AA_BP=2953 MAAAARPRGRALGPVLPPTPLLLLVLRVLPACGATARDPGAAAGLSLHPTYFNLAEAARIWATATCGERGPGEGRPQPELYCKLVGGPTA PGSGHTIQGQFCDYCNSEDPRKAHPVTNAIDGSERWWQSPPLSSGTQYNRVNLTLDLGQLFHVAYILIKFANSPRPDLWVLERSVDFGST YSPWQYFAHSKVDCLKEFGREANMAVTRDDDVLCVTEYSRIVPLENGEVVVSLINGRPGAKNFTFSHTLREFTKATNIRLRFLRTNTLLG HLISKAQRDPTVTRRYYYSIKDISIGGQCVCNGHAEVCNINNPEKLFRCECQHHTCGETCDRCCTGYNQRRWRPAAWEQSHECEACNCHG HASNCYYDPDVERQQASLNTQGIYAGGGVCINCQHNTAGVNCEQCAKGYYRPYGVPVDAPDGCIPCSCDPEHADGCEQGSGRCHCKPNFH GDNCEKCAIGYYNFPFCLRIPIFPVSTPSSEDPVAGDIKGCDCNLEGVLPEICDAHGRCLCRPGVEGPRCDTCRSGFYSFPICQACWCSA LGSYQMPCSSVTGQCECRPGVTGQRCDRCLSGAYDFPHCQGSSSACDPAGTINSNLGYCQCKLHVEGPTCSRCKLLYWNLDKENPSGCSE CKCHKAGTVSGTGECRQGDGDCHCKSHVGGDSCDTCEDGYFALEKSNYFGCQGCQCDIGGALSSMCSGPSGVCQCREHVVGKVCQRPENN YYFPDLHHMKYEIEDGSTPNGRDLRFGFDPLAFPEFSWRGYAQMTSVQNDVRITLNVGKSSGSLFRVILRYVNPGTEAVSGHITIYPSWG AAQSKEIIFLPSKEPAFVTVPGNGFADPFSITPGIWVACIKAEGVLLDYLVLLPRDYYEASVLQLPVTEPCAYAGPPQENCLLYQHLPVT RFPCTLACEARHFLLDGEPRPVAVRQPTPAHPVMVDLSGREVELHLRLRIPQVGHYVVVVEYSTEAAQLFVVDVNVKSSGSVLAGQVNIY SCNYSVLCRSAVIDHMSRIAMYELLADADIQLKGHMARFLLHQVCIIPIEEFSAEYVRPQVHCIASYGRFVNQSATCVSLAHETPPTALI LDVLSGRPFPHLPQQSSPSVDVLPGVTLKAPQNQVTLRGRVPHLGRYVFVIHFYQAAHPTFPAQVSVDGGWPRAGSFHASFCPHVLGCRD QVIAEGQIEFDISEPEVAATVKVPEGKSLVLVRVLVVPAENYDYQILHKKSMDKSLEFITNCGKNSFYLDPQTASRFCKNSARSLVAFYH KGALPCECHPTGATGPHCSPEGGQCPCQPNVIGRQCTRCATGHYGFPRCKPCSCGRRLCEEMTGQCRCPPRTVRPQCEVCETHSFSFHPM AGCEGCNCSRRGTIEAAMPECDRDSGQCRCKPRITGRQCDRCASGFYRFPECVPCNCNRDGTEPGVCDPGTGACLCKENVEGTECNVCRE GSFHLDPANLKGCTSCFCFGVNNQCHSSHKRRTKFVDMLGWHLETADRVDIPVSFNPGSNSMVADLQELPATIHSASWVAPTSYLGDKVS SYGGYLTYQAKSFGLPGDMVLLEKKPDVQLTGQHMSIIYEETNTPRPDRLHHGRVHVVEGNFRHASSRAPVSREELMTVLSRLADVRIQG LYFTETQRLTLSEVGLEEASDTGSGRIALAVEICACPPAYAGDSCQGCSPGYYRDHKGLYTGRCVPCNCNGHSNQCQDGSGICVNCQHNT AGEHCERCQEGYYGNAVHGSCRACPCPHTNSFATGCVVNGGDVRCSCKAGYTGTQCERCAPGYFGNPQKFGGSCQPCSCNSNGQLGSCHP LTGDCINQEPKDSSPAEECDDCDSCVMTLLNDLATMGEQLRLVKSQLQGLSASAGLLEQMRHMETQAKDLRNQLLNYRSAISNHGSKIEG LERELTDLNQEFETLQEKAQVNSRKAQTLNNNVNRATQSAKELDVKIKNVIRNVHILLKQISGTDGEGNNVPSGDFSREWAEAQRMMREL RNRNFGKHLREAEADKRESQLLLNRIRTWQKTHQGENNGLANSIRDSLNEYEAKLSDLRARLQEAAAQAKQANGLNQENERALGAIQRQV KEINSLQSDFTKYLTTADSSLLQTNIALQLMEKSQKEYEKLAASLNEARQELSDKVRELSRSAGKTSLVEEAEKHARSLQELAKQLEEIK RNASGDELVRCAVDAATAYENILNAIKAAEDAANRAASASESALQTVIKEDLPRKAKTLSSNSDKLLNEAKMTQKKLKQEVSPALNNLQQ TLNIVTVQKEVIDTNLTTLRDGLHGIQRGDIDAMISSAKSMVRKANDITDEVLDGLNPIQTDVERIKDTYGRTQNEDFKKALTDADNSVN KLTNKLPDLWRKIESINQQLLPLGNISDNMDRIRELIQQARDAASKVAVPMRFNGKSGVEVRLPNDLEDLKGYTSLSLFLQRPNSRENGG TENMFVMYLGNKDASRDYIGMAVVDGQLTCVYNLGDREAELQVDQILTKSETKEAVMDRVKFQRIYQFARLNYTKGATSSKPETPGVYDM DGRNSNTLLNLDPENVVFYVGGYPPDFKLPSRLSFPPYKGCIELDDLNENVLSLYNFKKTFNLNTTEVEPCRRRKEESDKNYFEGTGYAR VPTQPHAPIPTFGQTIQTTVDRGLLFFAENGDRFISLNIEDGKLMVRYKLNSELPKERGVGDAINNGRDHSIQIKIGKLQKRMWINVDVQ NTIIDGEVFDFSTYYLGGIPIAIRERFNISTPAFRGCMKNLKKTSGVVRLNDTVGVTKKCSEDWKLVRSASFSRGGQLSFTDLGLPPTDH LQASFGFQTFQPSGILLDHQTWTRNLQVTLEDGYIELSTSDSGSPIFKSPQTYMDGLLHYVSVISDNSGLRLLIDDQLLRNSKRLKHISS SRQSLRLGGSNFEGCISNVFVQRLSLSPEVLDLTSNSLKRDVSLGGCSLNKPPFLMLLKGSTRFNKTKTFRINQDWLEAIEEHSAYSTHY CSQDQLTDEEEEDTVSAADLKKSLEKAQSCQQRLDREISNFLKMIKECDMAKEMLPSFLQKVEVVSEASRETCVALTDCLNLFTKQEGVR NFKLEQEQEKNKILSEALETLATEHHELEQSLVKGSPPASILSEDEFYDALSDSESERSLSRLEAVTARSFEEEGEHLGSRKHRMSEEKD CGGGDALSNGIKKHRTSLPSPMFSRNDFSIWSILRKCIGMELSKITMPVIFNEPLSFLQRLTEYMEHTYLIHKASSLSDPVERMQCVAAF AVSAVASQWERTGKPFNPLLGETYELVRDDLGFRLISEQVSHHPPISAFHAEGLNNDFIFHGSIYPKLKFWGKSVEAEPKGTITLELLEH NEAYTWTNPTCCVHNIIVGKLWIEQYGNVEIINHKTGDKCVLNFKPCGLFGKELHKVEGYIQDKSKKKLCALYGKWTECLYSVDPATFDA YKKNDKKNTEEKKNSKQMSTSEELDEMPVPDSESVFIIPGSVLLWRIAPRPPNSAQMYNFTSFAMVLNEVDKDMESVIPKTDCRLRPDIR -------------------------------------------------------------- >43877_43877_3_LAMA3-OSBPL1A_LAMA3_chr18_21513899_ENST00000399516_OSBPL1A_chr18_21892070_ENST00000319481_length(amino acids)=3525AA_BP=2897 MAAAARPRGRALGPVLPPTPLLLLVLRVLPACGATARDPGAAAGLSLHPTYFNLAEAARIWATATCGERGPGEGRPQPELYCKLVGGPTA PGSGHTIQGQFCDYCNSEDPRKAHPVTNAIDGSERWWQSPPLSSGTQYNRVNLTLDLGQLFHVAYILIKFANSPRPDLWVLERSVDFGST YSPWQYFAHSKVDCLKEFGREANMAVTRDDDVLCVTEYSRIVPLENGEVVVSLINGRPGAKNFTFSHTLREFTKATNIRLRFLRTNTLLG HLISKAQRDPTVTRRYYYSIKDISIGGQCVCNGHAEVCNINNPEKLFRCECQHHTCGETCDRCCTGYNQRRWRPAAWEQSHECEACNCHG HASNCYYDPDVERQQASLNTQGIYAGGGVCINCQHNTAGVNCEQCAKGYYRPYGVPVDAPDGCIPCSCDPEHADGCEQGSGRCHCKPNFH GDNCEKCAIGYYNFPFCLRIPIFPVSTPSSEDPVAGDIKGCDCNLEGVLPEICDAHGRCLCRPGVEGPRCDTCRSGFYSFPICQACWCSA LGSYQMPCSSVTGQCECRPGVTGQRCDRCLSGAYDFPHCQGSSSACDPAGTINSNLGYCQCKLHVEGPTCSRCKLLYWNLDKENPSGCSE CKCHKAGTVSGTGECRQGDGDCHCKSHVGGDSCDTCEDGYFALEKSNYFGCQGCQCDIGGALSSMCSGPSGVCQCREHVVGKVCQRPENN YYFPDLHHMKYEIEDGSTPNGRDLRFGFDPLAFPEFSWRGYAQMTSVQNDVRITLNVGKSSGSLFRVILRYVNPGTEAVSGHITIYPSWG AAQSKEIIFLPSKEPAFVTVPGNGFADPFSITPGIWVACIKAEGVLLDYLVLLPRDYYEASVLQLPVTEPCAYAGPPQENCLLYQHLPVT RFPCTLACEARHFLLDGEPRPVAVRQPTPAHPVMVDLSGREVELHLRLRIPQVGHYVVVVEYSTEAAQLFVVDVNVKSSGSVLAGQVNIY SCNYSVLCRSAVIDHMSRIAMYELLADADIQLKGHMARFLLHQVCIIPIEEFSAEYVRPQVHCIASYGRFVNQSATCVSLAHETPPTALI LDVLSGRPFPHLPQQSSPSVDVLPGVTLKAPQNQVTLRGRVPHLGRYVFVIHFYQAAHPTFPAQVSVDGGWPRAGSFHASFCPHVLGCRD QVIAEGQIEFDISEPEVAATVKVPEGKSLVLVRVLVVPAENYDYQILHKKSMDKSLEFITNCGKNSFYLDPQTASRFCKNSARSLVAFYH KGALPCECHPTGATGPHCSPEGGQCPCQPNVIGRQCTRCATGHYGFPRCKPCSCGRRLCEEMTGQCRCPPRTVRPQCEVCETHSFSFHPM AGCEGCNCSRRGTIEAAMPECDRDSGQCRCKPRITGRQCDRCASGFYRFPECVPCNCNRDGTEPGVCDPGTGACLCKENVEGTECNVCRE GSFHLDPANLKGCTSCFCFGVNNQCHSSHKRRTKFVDMLGWHLETADRVDIPVSFNPGSNSMVADLQELPATIHSASWVAPTSYLGDKVS SYGGYLTYQAKSFGLPGDMVLLEKKPDVQLTGQHMSIIYEETNTPRPDRLHHGRVHVVEGNFRHASSRAPVSREELMTVLSRLADVRIQG LYFTETQRLTLSEVGLEEASDTGSGRIALAVEICACPPAYAGDSCQGCSPGYYRDHKGLYTGRCVPCNCNGHSNQCQDGSGICVNCQHNT AGEHCERCQEGYYGNAVHGSCRACPCPHTNSFATGCVVNGGDVRCSCKAGYTGTQCERCAPGYFGNPQKFGGSCQPCSCNSNGQLGSCHP LTGDCINQEPKDSSPAEECDDCDSCVMTLLNDLATMGEQLRLVKSQLQGLSASAGLLEQMRHMETQAKDLRNQLLNYRSAISNHGSKIEG LERELTDLNQEFETLQEKAQVNSRKAQTLNNNVNRATQSAKELDVKIKNVIRNVHMLNRIRTWQKTHQGENNGLANSIRDSLNEYEAKLS DLRARLQEAAAQAKQANGLNQENERALGAIQRQVKEINSLQSDFTKYLTTADSSLLQTNIALQLMEKSQKEYEKLAASLNEARQELSDKV RELSRSAGKTSLVEEAEKHARSLQELAKQLEEIKRNASGDELVRCAVDAATAYENILNAIKAAEDAANRAASASESALQTVIKEDLPRKA KTLSSNSDKLLNEAKMTQKKLKQEVSPALNNLQQTLNIVTVQKEVIDTNLTTLRDGLHGIQRGDIDAMISSAKSMVRKANDITDEVLDGL NPIQTDVERIKDTYGRTQNEDFKKALTDADNSVNKLTNKLPDLWRKIESINQQLLPLGNISDNMDRIRELIQQARDAASKVAVPMRFNGK SGVEVRLPNDLEDLKGYTSLSLFLQRPNSRENGGTENMFVMYLGNKDASRDYIGMAVVDGQLTCVYNLGDREAELQVDQILTKSETKEAV MDRVKFQRIYQFARLNYTKGATSSKPETPGVYDMDGRNSNTLLNLDPENVVFYVGGYPPDFKLPSRLSFPPYKGCIELDDLNENVLSLYN FKKTFNLNTTEVEPCRRRKEESDKNYFEGTGYARVPTQPHAPIPTFGQTIQTTVDRGLLFFAENGDRFISLNIEDGKLMVRYKLNSELPK ERGVGDAINNGRDHSIQIKIGKLQKRMWINVDVQNTIIDGEVFDFSTYYLGGIPIAIRERFNISTPAFRGCMKNLKKTSGVVRLNDTVGV TKKCSEDWKLVRSASFSRGGQLSFTDLGLPPTDHLQASFGFQTFQPSGILLDHQTWTRNLQVTLEDGYIELSTSDSGSPIFKSPQTYMDG LLHYVSVISDNSGLRLLIDDQLLRNSKRLKHISSSRQSLRLGGSNFEGCISNVFVQRLSLSPEVLDLTSNSLKRDVSLGGCSLNKPPFLM LLKGSTRFNKTKTFRINQDWLEAIEEHSAYSTHYCSQDQLTDEEEEDTVSAADLKKSLEKAQSCQQRLDREISNFLKMIKECDMAKEMLP SFLQKVEVVSEASRETCVALTDCLNLFTKQEGVRNFKLEQEQEKNKILSEALETLATEHHELEQSLVKGSPPASILSEDEFYDALSDSES ERSLSRLEAVTARSFEEEGEHLGSRKHRMSEEKDCGGGDALSNGIKKHRTSLPSPMFSRNDFSIWSILRKCIGMELSKITMPVIFNEPLS FLQRLTEYMEHTYLIHKASSLSDPVERMQCVAAFAVSAVASQWERTGKPFNPLLGETYELVRDDLGFRLISEQVSHHPPISAFHAEGLNN DFIFHGSIYPKLKFWGKSVEAEPKGTITLELLEHNEAYTWTNPTCCVHNIIVGKLWIEQYGNVEIINHKTGDKCVLNFKPCGLFGKELHK VEGYIQDKSKKKLCALYGKWTECLYSVDPATFDAYKKNDKKNTEEKKNSKQMSTSEELDEMPVPDSESVFIIPGSVLLWRIAPRPPNSAQ MYNFTSFAMVLNEVDKDMESVIPKTDCRLRPDIRAMENGEIDQASEEKKRLEEKQRAARKNRSKSEEDWKTRWFHQGPNPYNGAQDWIYS -------------------------------------------------------------- >43877_43877_4_LAMA3-OSBPL1A_LAMA3_chr18_21513899_ENST00000587184_OSBPL1A_chr18_21892070_ENST00000319481_length(amino acids)=1916AA_BP=1288 MPPAVRRSACSMGWLWIFGAALGQCLGYSSQQQRVPFLQPPGQSQLQASYVEFRPSQGCSPGYYRDHKGLYTGRCVPCNCNGHSNQCQDG SGICVNCQHNTAGEHCERCQEGYYGNAVHGSCRACPCPHTNSFATGCVVNGGDVRCSCKAGYTGTQCERCAPGYFGNPQKFGGSCQPCSC NSNGQLGSCHPLTGDCINQEPKDSSPAEECDDCDSCVMTLLNDLATMGEQLRLVKSQLQGLSASAGLLEQMRHMETQAKDLRNQLLNYRS AISNHGSKIEGLERELTDLNQEFETLQEKAQVNSRKAQTLNNNVNRATQSAKELDVKIKNVIRNVHMLNRIRTWQKTHQGENNGLANSIR DSLNEYEAKLSDLRARLQEAAAQAKQANGLNQENERALGAIQRQVKEINSLQSDFTKYLTTADSSLLQTNIALQLMEKSQKEYEKLAASL NEARQELSDKVRELSRSAGKTSLVEEAEKHARSLQELAKQLEEIKRNASGDELVRCAVDAATAYENILNAIKAAEDAANRAASASESALQ TVIKEDLPRKAKTLSSNSDKLLNEAKMTQKKLKQEVSPALNNLQQTLNIVTVQKEVIDTNLTTLRDGLHGIQRGDIDAMISSAKSMVRKA NDITDEVLDGLNPIQTDVERIKDTYGRTQNEDFKKALTDADNSVNKLTNKLPDLWRKIESINQQLLPLGNISDNMDRIRELIQQARDAAS KVAVPMRFNGKSGVEVRLPNDLEDLKGYTSLSLFLQRPNSRENGGTENMFVMYLGNKDASRDYIGMAVVDGQLTCVYNLGDREAELQVDQ ILTKSETKEAVMDRVKFQRIYQFARLNYTKGATSSKPETPGVYDMDGRNSNTLLNLDPENVVFYVGGYPPDFKLPSRLSFPPYKGCIELD DLNENVLSLYNFKKTFNLNTTEVEPCRRRKEESDKNYFEGTGYARVPTQPHAPIPTFGQTIQTTVDRGLLFFAENGDRFISLNIEDGKLM VRYKLNSELPKERGVGDAINNGRDHSIQIKIGKLQKRMWINVDVQNTIIDGEVFDFSTYYLGGIPIAIRERFNISTPAFRGCMKNLKKTS GVVRLNDTVGVTKKCSEDWKLVRSASFSRGGQLSFTDLGLPPTDHLQASFGFQTFQPSGILLDHQTWTRNLQVTLEDGYIELSTSDSGSP IFKSPQTYMDGLLHYVSVISDNSGLRLLIDDQLLRNSKRLKHISSSRQSLRLGGSNFEGCISNVFVQRLSLSPEVLDLTSNSLKRDVSLG GCSLNKPPFLMLLKGSTRFNKTKTFRINQDWLEAIEEHSAYSTHYCSQDQLTDEEEEDTVSAADLKKSLEKAQSCQQRLDREISNFLKMI KECDMAKEMLPSFLQKVEVVSEASRETCVALTDCLNLFTKQEGVRNFKLEQEQEKNKILSEALETLATEHHELEQSLVKGSPPASILSED EFYDALSDSESERSLSRLEAVTARSFEEEGEHLGSRKHRMSEEKDCGGGDALSNGIKKHRTSLPSPMFSRNDFSIWSILRKCIGMELSKI TMPVIFNEPLSFLQRLTEYMEHTYLIHKASSLSDPVERMQCVAAFAVSAVASQWERTGKPFNPLLGETYELVRDDLGFRLISEQVSHHPP ISAFHAEGLNNDFIFHGSIYPKLKFWGKSVEAEPKGTITLELLEHNEAYTWTNPTCCVHNIIVGKLWIEQYGNVEIINHKTGDKCVLNFK PCGLFGKELHKVEGYIQDKSKKKLCALYGKWTECLYSVDPATFDAYKKNDKKNTEEKKNSKQMSTSEELDEMPVPDSESVFIIPGSVLLW RIAPRPPNSAQMYNFTSFAMVLNEVDKDMESVIPKTDCRLRPDIRAMENGEIDQASEEKKRLEEKQRAARKNRSKSEEDWKTRWFHQGPN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:21513899/chr18:21892070) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LAMA3 | . |
| FUNCTION: Binding to cells via a high affinity receptor, laminin is thought to mediate the attachment, migration and organization of cells into tissues during embryonic development by interacting with other extracellular matrix components.; FUNCTION: Laminin-5 is thought to be involved in (1) cell adhesion via integrin alpha-3/beta-1 in focal adhesion and integrin alpha-6/beta-4 in hemidesmosomes, (2) signal transduction via tyrosine phosphorylation of pp125-FAK and p80, (3) differentiation of keratinocytes. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1852_1941 | 2954.0 | 3334.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1987_2169 | 2954.0 | 3334.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2322_2388 | 2954.0 | 3334.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1852_1941 | 2898.0 | 3278.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1987_2169 | 2898.0 | 3278.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2322_2388 | 2898.0 | 3278.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1266_1311 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 9 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1312_1355 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 10 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1356_1404 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 11 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1405_1455 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 12 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1476_1653 | 2954.0 | 3334.0 | Domain | Laminin IV type A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1687_1733 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 13 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1734_1786 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 14 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1787_1821 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 15%3B truncated |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2390_2591 | 2954.0 | 3334.0 | Domain | Laminin G-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2598_2760 | 2954.0 | 3334.0 | Domain | Laminin G-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2767_2927 | 2954.0 | 3334.0 | Domain | Laminin G-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 299_355 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 356_425 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 426_469 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 43_298 | 2954.0 | 3334.0 | Domain | Laminin N-terminal |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 491_535 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 536_588 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 590_630 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 6 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 631_683 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 7 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 684_728 | 2954.0 | 3334.0 | Domain | Laminin EGF-like 8 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1266_1311 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 9 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1312_1355 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 10 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1356_1404 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 11 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1405_1455 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 12 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1476_1653 | 2898.0 | 3278.0 | Domain | Laminin IV type A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1687_1733 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 13 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1734_1786 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 14 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1787_1821 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 15%3B truncated |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2390_2591 | 2898.0 | 3278.0 | Domain | Laminin G-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2598_2760 | 2898.0 | 3278.0 | Domain | Laminin G-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 299_355 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 356_425 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 426_469 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 43_298 | 2898.0 | 3278.0 | Domain | Laminin N-terminal |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 491_535 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 536_588 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 590_630 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 6 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 631_683 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 7 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 684_728 | 2898.0 | 3278.0 | Domain | Laminin EGF-like 8 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 299_355 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 356_425 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 426_469 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 43_298 | 1289.0 | 1669.0 | Domain | Laminin N-terminal |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 491_535 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 536_588 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 590_630 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 6 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 631_683 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 7 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 684_728 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 8 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2278_2280 | 2954.0 | 3334.0 | Motif | Cell attachment site |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2278_2280 | 2898.0 | 3278.0 | Motif | Cell attachment site |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1266_1465 | 2954.0 | 3334.0 | Region | Note=Domain III B |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1654_1821 | 2954.0 | 3334.0 | Region | Note=Domain III A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 1822_2389 | 2954.0 | 3334.0 | Region | Note=Domain II and I |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 298_728 | 2954.0 | 3334.0 | Region | Note=Domain V |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 796_1265 | 2954.0 | 3334.0 | Region | Note=Domain IV 1 (domain IV B) |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1266_1465 | 2898.0 | 3278.0 | Region | Note=Domain III B |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1654_1821 | 2898.0 | 3278.0 | Region | Note=Domain III A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 1822_2389 | 2898.0 | 3278.0 | Region | Note=Domain II and I |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 298_728 | 2898.0 | 3278.0 | Region | Note=Domain V |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 796_1265 | 2898.0 | 3278.0 | Region | Note=Domain IV 1 (domain IV B) |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 298_728 | 1289.0 | 1669.0 | Region | Note=Domain V |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 796_1265 | 1289.0 | 1669.0 | Region | Note=Domain IV 1 (domain IV B) |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 430_463 | 323.0 | 951.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 877_913 | 323.0 | 951.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 430_463 | 0 | 569.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 877_913 | 0 | 569.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 430_463 | 0 | 438.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 877_913 | 0 | 438.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 235_334 | 0 | 569.0 | Domain | PH | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 235_334 | 0 | 438.0 | Domain | PH | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 469_483 | 323.0 | 951.0 | Motif | FFAT | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 469_483 | 0 | 569.0 | Motif | FFAT | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 469_483 | 0 | 438.0 | Motif | FFAT | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 175_204 | 0 | 569.0 | Repeat | Note=ANK 3 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 47_76 | 0 | 569.0 | Repeat | Note=ANK 1 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000357041 | 0 | 16 | 80_109 | 0 | 569.0 | Repeat | Note=ANK 2 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 175_204 | 0 | 438.0 | Repeat | Note=ANK 3 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 47_76 | 0 | 438.0 | Repeat | Note=ANK 1 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000399443 | 0 | 14 | 80_109 | 0 | 438.0 | Repeat | Note=ANK 2 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1852_1941 | 1289.0 | 1669.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1987_2169 | 1289.0 | 1669.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2322_2388 | 1289.0 | 1669.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 2986_3150 | 2954.0 | 3334.0 | Domain | Laminin G-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000313654 | + | 67 | 75 | 3157_3330 | 2954.0 | 3334.0 | Domain | Laminin G-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2767_2927 | 2898.0 | 3278.0 | Domain | Laminin G-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 2986_3150 | 2898.0 | 3278.0 | Domain | Laminin G-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000399516 | + | 66 | 74 | 3157_3330 | 2898.0 | 3278.0 | Domain | Laminin G-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1266_1311 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 9 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1312_1355 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 10 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1356_1404 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 11 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1405_1455 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 12 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1476_1653 | 1289.0 | 1669.0 | Domain | Laminin IV type A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1687_1733 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 13 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1734_1786 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 14 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1787_1821 | 1289.0 | 1669.0 | Domain | Laminin EGF-like 15%3B truncated |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2390_2591 | 1289.0 | 1669.0 | Domain | Laminin G-like 1 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2598_2760 | 1289.0 | 1669.0 | Domain | Laminin G-like 2 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2767_2927 | 1289.0 | 1669.0 | Domain | Laminin G-like 3 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2986_3150 | 1289.0 | 1669.0 | Domain | Laminin G-like 4 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 3157_3330 | 1289.0 | 1669.0 | Domain | Laminin G-like 5 |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 2278_2280 | 1289.0 | 1669.0 | Motif | Cell attachment site |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1266_1465 | 1289.0 | 1669.0 | Region | Note=Domain III B |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1654_1821 | 1289.0 | 1669.0 | Region | Note=Domain III A |
| Hgene | LAMA3 | chr18:21513899 | chr18:21892070 | ENST00000587184 | + | 29 | 37 | 1822_2389 | 1289.0 | 1669.0 | Region | Note=Domain II and I |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 235_334 | 323.0 | 951.0 | Domain | PH | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 175_204 | 323.0 | 951.0 | Repeat | Note=ANK 3 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 47_76 | 323.0 | 951.0 | Repeat | Note=ANK 1 | |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 80_109 | 323.0 | 951.0 | Repeat | Note=ANK 2 |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
LAMA3_pLDDT.png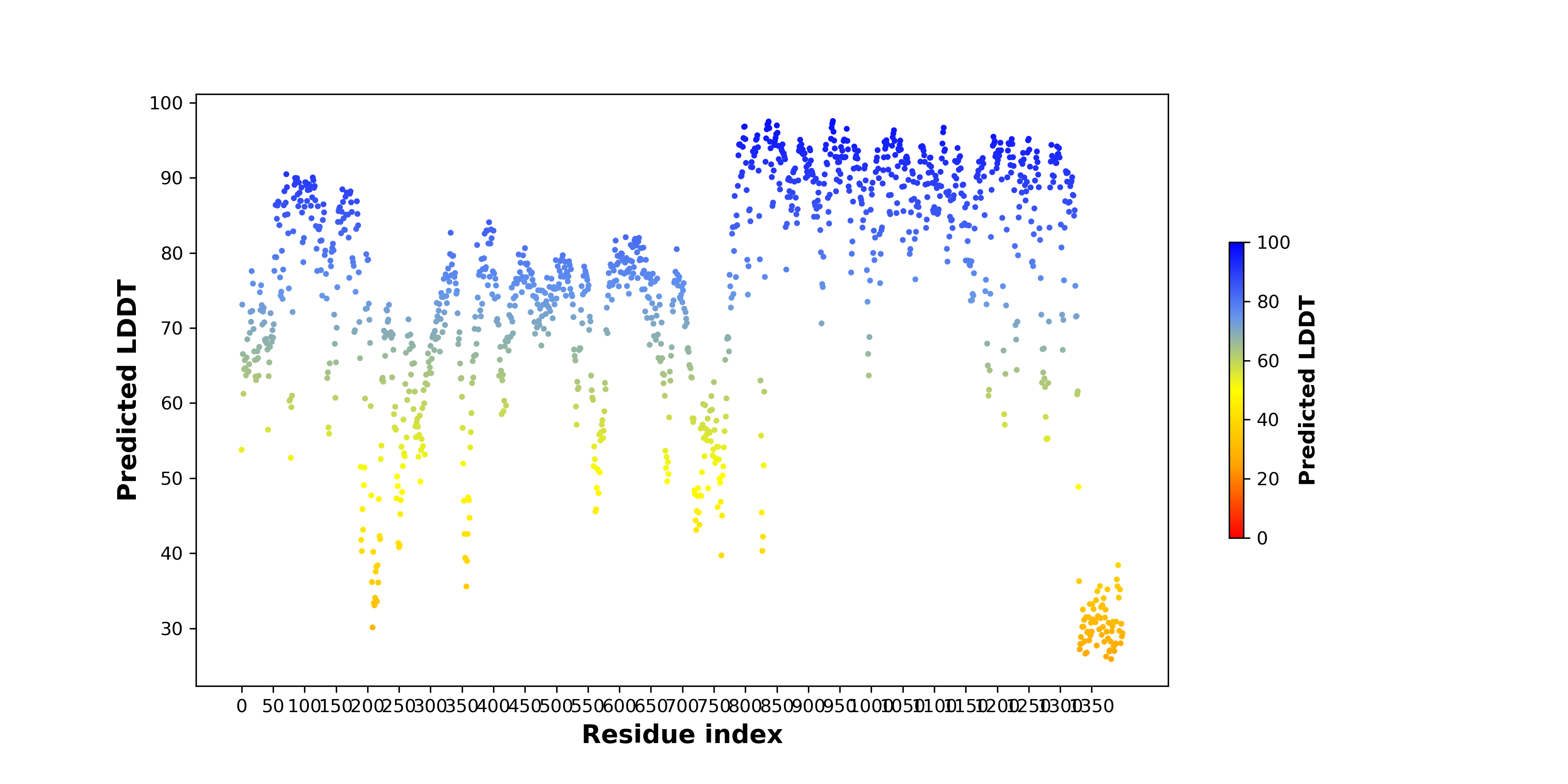 |
OSBPL1A_pLDDT.png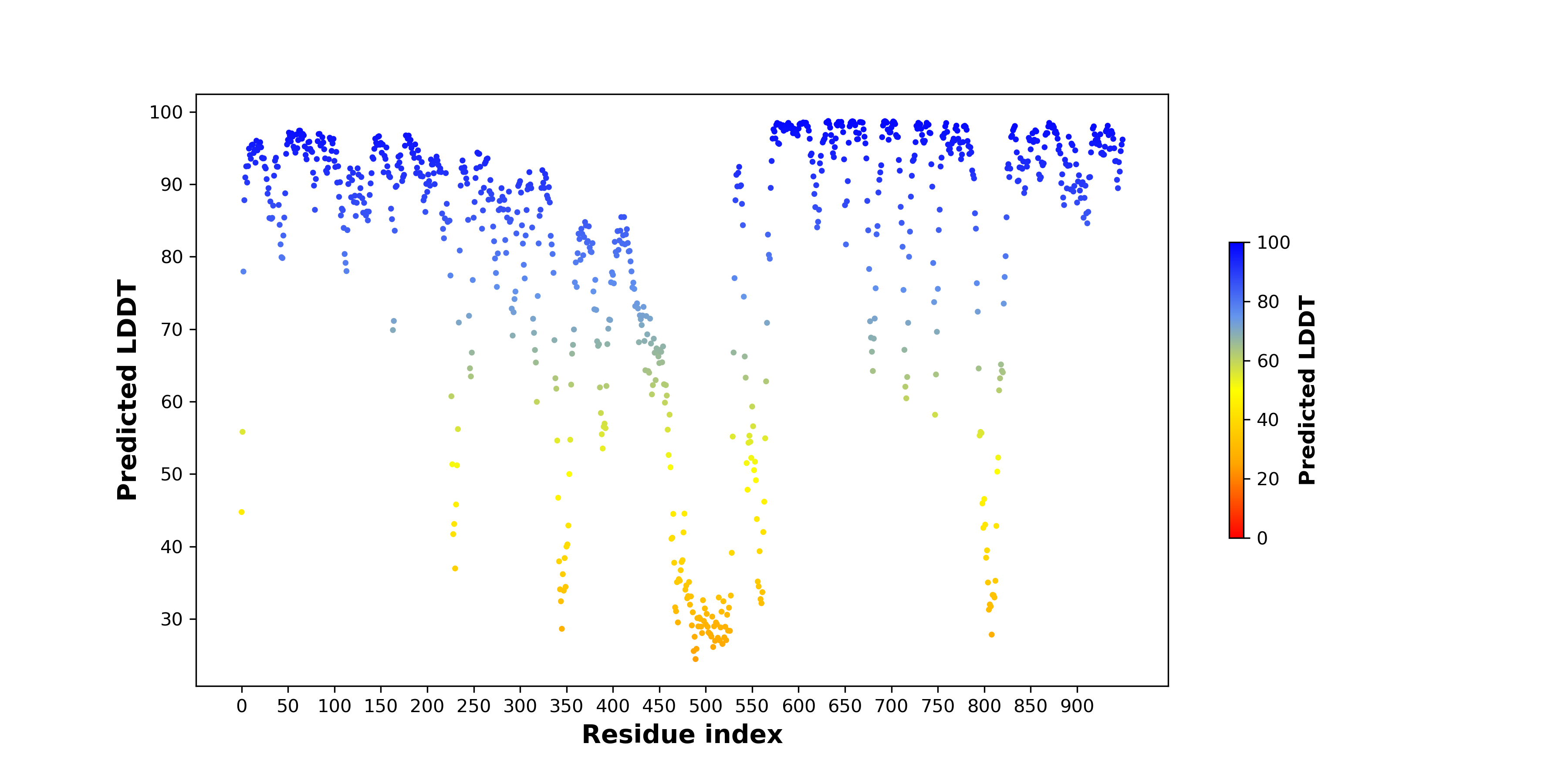 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| LAMA3 | |
| OSBPL1A |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | OSBPL1A | chr18:21513899 | chr18:21892070 | ENST00000319481 | 11 | 28 | 1_237 | 323.0 | 951.0 | RAB7A |
Top |
Related Drugs to LAMA3-OSBPL1A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to LAMA3-OSBPL1A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | LAMA3 | C1328355 | Laryngoonychocutaneous syndrome | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | LAMA3 | C0079683 | Herlitz Disease | 4 | CTD_human;GENOMICS_ENGLAND |
| Hgene | LAMA3 | C0079301 | Junctional Epidermolysis Bullosa | 3 | CTD_human;GENOMICS_ENGLAND |
| Hgene | LAMA3 | C0268374 | Adult junctional epidermolysis bullosa (disorder) | 3 | CTD_human;GENOMICS_ENGLAND |
| Hgene | LAMA3 | C0027339 | Nail Diseases | 1 | CTD_human |
| Hgene | LAMA3 | C0037299 | Skin Ulcer | 1 | CTD_human |
| Hgene | LAMA3 | C0079297 | Epidermolysis Bullosa Progressiva | 1 | CTD_human |
| Hgene | LAMA3 | C0399367 | Amelogenesis imperfecta local hypoplastic form | 1 | GENOMICS_ENGLAND |

