| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MAPKAPK3-DOCK3 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MAPKAPK3-DOCK3 | FusionPDB ID: 51615 | FusionGDB2.0 ID: 51615 | Hgene | Tgene | Gene symbol | MAPKAPK3 | DOCK3 | Gene ID | 7867 | 1795 |
| Gene name | MAPK activated protein kinase 3 | dedicator of cytokinesis 3 | |
| Synonyms | 3PK|MAPKAP-K3|MAPKAP3|MAPKAPK-3|MDPT3|MK-3|MK3 | MOCA|NEDIDHA|PBP | |
| Cytomap | 3p21.2 | 3p21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | MAP kinase-activated protein kinase 3MAPKAP kinase 3chromosome 3p kinasemitogen-activated protein kinase-activated protein kinase 3 | dedicator of cytokinesis protein 3modifier of cell adhesionpresenilin-binding protein | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q16644 | Q8IZD9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000357955, ENST00000446044, ENST00000497283, | ENST00000266037, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 1 X 3=9 | 12 X 10 X 7=840 |
| # samples | 3 | 12 | |
| ** MAII score | log2(3/9*10)=1.73696559416621 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(12/840*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MAPKAPK3 [Title/Abstract] AND DOCK3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MAPKAPK3(50655215)-DOCK3(50927457), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MAPKAPK3-DOCK3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MAPKAPK3-DOCK3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MAPKAPK3-DOCK3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MAPKAPK3-DOCK3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MAPKAPK3 | GO:0018105 | peptidyl-serine phosphorylation | 15850461 |
| Hgene | MAPKAPK3 | GO:0034097 | response to cytokine | 8774846 |
 Fusion gene breakpoints across MAPKAPK3 (5'-gene) Fusion gene breakpoints across MAPKAPK3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
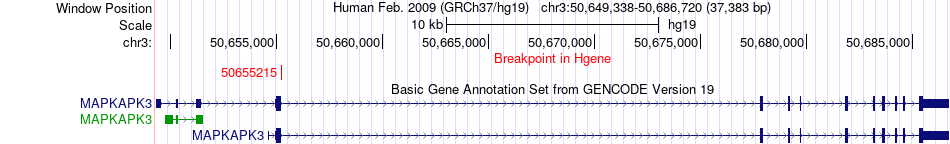 |
 Fusion gene breakpoints across DOCK3 (3'-gene) Fusion gene breakpoints across DOCK3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
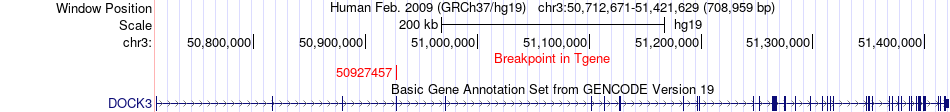 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-58-A46L | MAPKAPK3 | chr3 | 50655215 | + | DOCK3 | chr3 | 50927457 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000446044 | MAPKAPK3 | chr3 | 50655215 | + | ENST00000266037 | DOCK3 | chr3 | 50927457 | + | 9385 | 815 | 596 | 6745 | 2049 |
| ENST00000357955 | MAPKAPK3 | chr3 | 50655215 | + | ENST00000266037 | DOCK3 | chr3 | 50927457 | + | 8898 | 328 | 109 | 6258 | 2049 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000446044 | ENST00000266037 | MAPKAPK3 | chr3 | 50655215 | + | DOCK3 | chr3 | 50927457 | + | 0.000959173 | 0.9990408 |
| ENST00000357955 | ENST00000266037 | MAPKAPK3 | chr3 | 50655215 | + | DOCK3 | chr3 | 50927457 | + | 0.000645556 | 0.9993544 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >51615_51615_1_MAPKAPK3-DOCK3_MAPKAPK3_chr3_50655215_ENST00000357955_DOCK3_chr3_50927457_ENST00000266037_length(amino acids)=2049AA_BP=0 MDGETAEEQGGPVPPPVAPGGPGLGGAPGGRREPKKYAVTDDYQLSKQVLGLGVNGKVLECFHRRTGQKCALKGIFPANYIHLKKAIVSN RGQYETVVPLEDSIVTEVTATLQEWASLWKQLYVKHKVDLFYKLRHVMNELIDLRRQLLSGHLTQDQVREVKRHITVRLDWGNEHLGLDL VPRKDFEVVDSDQISVSDLYKMHLSSRQSVQQSTSQVDTMRPRHGETCRMPVPHHFFLSLKSFTYNTIGEDTDVFFSLYDMREGKQISER FLVRLNKNGGPRNPEKIERMCALFTDLSSKDMKRDLYIVAHVIRIGRMLLNDSKKGPPHLHYRRPYGCAVLSILDVLQSLTEVKEEKDFV LKVYTCNNESEWSQIHENIIRKSSAKYSAPSASHGLIISLQLLRGDMEQIRRENPMIFNRGLAITRKLGFPDVIMPGDIRNDLYLTLEKG DFERGGKSVQKNIEVTMYVLYADGEILKDCISLGSGEPNRSSYHSFVLYHSNSPRWGEIIKLPIPIDRFRGSHLRFEFRHCSTKDKGEKK LFGFAFSTLMRDDGTTLSDDIHELYVYKCDENSTFNNHALYLGLPCCKEDYNGCPNIPSSLIFQRSTKESFFISTQLSSTKLTQNVDLLA LLKWKAFPDRIMDVLGRLRHVSGEEIVKFLQDILDTLFVILDDNTEKYGLLVFQSLVFIINLLRDIKYFHFRPVMDTYIQKHFAGALAYK ELIRCLKWYMDCSAELIRQDHIQEAMRALEYLFKFIVQSRILYSRATCGMEEEQFRSSIQELFQSIRFVLSLDSRNSETLLFTQAALLNS FPTIFDELLQMFTVQEVAEFVRGTLGSMPSTVHIGQSMDVVKLQSIARTVDSRLFSFSESRRILLPVVLHHIHLHLRQQKELLICSGILG SIFSIVKTSSLEADVMEEVEMMVESLLDVLLQTLLTIMSKSHAQEAVRGQRCPQCTAEITGEYVSCLLSLLRQMCDTHFQHLLDNFQSKD ELKEFLLKIFCVFRNLMKMSVFPRDWMVMRLLTSNIIVTTVQYLSSALHKNFTETDFDFKVWNSYFSLAVLFINQPSLQLEIITSAKRKK ILDKYGDMRVMMAYELFSMWQNLGEHKIHFIPGMIGPFLGVTLVPQPEVRNIMIPIFHDMMDWEQRKNGNFKQVEAELIDKLDSMVSEGK GDESYRELFSLLTQLFGPYPSLLEKVEQETWRETGISFVTSVTRLMERLLDYRDCMKGEETENKKIGCTVNLMNFYKSEINKEEMYIRYI HKLCDMHLQAENYTEAAFTLLLYCELLQWEDRPLREFLHYPSQTEWQRKEGLCRKIIHYFNKGKSWEFGIPLCRELACQYESLYDYQSLS WIRKMEASYYDNIMEQQRLEPEFFRVGFYGRKFPFFLRNKEYVCRGHDYERLEAFQQRMLSEFPQAVAMQHPNHPDDAILQCDAQYLQIY AVTPIPDYVDVLQMDRVPDRVKSFYRVNNVRKFRYDRPFHKGPKDKENEFKSLWIERTTLTLTHSLPGISRWFEVERRELVEVSPLENAI QVVENKNQELRSLISQYQHKQVHGNINLLSMCLNGVIDAAVNGGIARYQEAFFDKDYINKHPGDAEKITQLKELMQEQVHVLGVGLAVHE KFVHPEMRPLHKKLIDQFQMMRASLYHEFPGLDKLSPACSGTSTPRGNVLASHSPMSPESIKMTHRHSPMNLMGTGRHSSSSLSSHASSE AGNMVMLGDGSMGDAPEDLYHHMQLAYPNPRYQGSVTNVSVLSSSQASPSSSSLSSTHSAPSQMITSAPSSARGSPSLPDKYRHAREMML LLPTYRDRPSSAMYPAAILENGQPPNFQRALFQQVVGACKPCSDPNLSVAEKGHYSLHFDAFHHPLGDTPPALPARTLRKSPLHPIPASP TSPQSGLDGSNSTLSGSASSGVSSLSESNFGHSSEAPPRTDTMDSMPSQAWNADEDLEPPYLPVHYSLSESAVLDSIKAQPCRSHSAPGC -------------------------------------------------------------- >51615_51615_2_MAPKAPK3-DOCK3_MAPKAPK3_chr3_50655215_ENST00000446044_DOCK3_chr3_50927457_ENST00000266037_length(amino acids)=2049AA_BP=0 MDGETAEEQGGPVPPPVAPGGPGLGGAPGGRREPKKYAVTDDYQLSKQVLGLGVNGKVLECFHRRTGQKCALKGIFPANYIHLKKAIVSN RGQYETVVPLEDSIVTEVTATLQEWASLWKQLYVKHKVDLFYKLRHVMNELIDLRRQLLSGHLTQDQVREVKRHITVRLDWGNEHLGLDL VPRKDFEVVDSDQISVSDLYKMHLSSRQSVQQSTSQVDTMRPRHGETCRMPVPHHFFLSLKSFTYNTIGEDTDVFFSLYDMREGKQISER FLVRLNKNGGPRNPEKIERMCALFTDLSSKDMKRDLYIVAHVIRIGRMLLNDSKKGPPHLHYRRPYGCAVLSILDVLQSLTEVKEEKDFV LKVYTCNNESEWSQIHENIIRKSSAKYSAPSASHGLIISLQLLRGDMEQIRRENPMIFNRGLAITRKLGFPDVIMPGDIRNDLYLTLEKG DFERGGKSVQKNIEVTMYVLYADGEILKDCISLGSGEPNRSSYHSFVLYHSNSPRWGEIIKLPIPIDRFRGSHLRFEFRHCSTKDKGEKK LFGFAFSTLMRDDGTTLSDDIHELYVYKCDENSTFNNHALYLGLPCCKEDYNGCPNIPSSLIFQRSTKESFFISTQLSSTKLTQNVDLLA LLKWKAFPDRIMDVLGRLRHVSGEEIVKFLQDILDTLFVILDDNTEKYGLLVFQSLVFIINLLRDIKYFHFRPVMDTYIQKHFAGALAYK ELIRCLKWYMDCSAELIRQDHIQEAMRALEYLFKFIVQSRILYSRATCGMEEEQFRSSIQELFQSIRFVLSLDSRNSETLLFTQAALLNS FPTIFDELLQMFTVQEVAEFVRGTLGSMPSTVHIGQSMDVVKLQSIARTVDSRLFSFSESRRILLPVVLHHIHLHLRQQKELLICSGILG SIFSIVKTSSLEADVMEEVEMMVESLLDVLLQTLLTIMSKSHAQEAVRGQRCPQCTAEITGEYVSCLLSLLRQMCDTHFQHLLDNFQSKD ELKEFLLKIFCVFRNLMKMSVFPRDWMVMRLLTSNIIVTTVQYLSSALHKNFTETDFDFKVWNSYFSLAVLFINQPSLQLEIITSAKRKK ILDKYGDMRVMMAYELFSMWQNLGEHKIHFIPGMIGPFLGVTLVPQPEVRNIMIPIFHDMMDWEQRKNGNFKQVEAELIDKLDSMVSEGK GDESYRELFSLLTQLFGPYPSLLEKVEQETWRETGISFVTSVTRLMERLLDYRDCMKGEETENKKIGCTVNLMNFYKSEINKEEMYIRYI HKLCDMHLQAENYTEAAFTLLLYCELLQWEDRPLREFLHYPSQTEWQRKEGLCRKIIHYFNKGKSWEFGIPLCRELACQYESLYDYQSLS WIRKMEASYYDNIMEQQRLEPEFFRVGFYGRKFPFFLRNKEYVCRGHDYERLEAFQQRMLSEFPQAVAMQHPNHPDDAILQCDAQYLQIY AVTPIPDYVDVLQMDRVPDRVKSFYRVNNVRKFRYDRPFHKGPKDKENEFKSLWIERTTLTLTHSLPGISRWFEVERRELVEVSPLENAI QVVENKNQELRSLISQYQHKQVHGNINLLSMCLNGVIDAAVNGGIARYQEAFFDKDYINKHPGDAEKITQLKELMQEQVHVLGVGLAVHE KFVHPEMRPLHKKLIDQFQMMRASLYHEFPGLDKLSPACSGTSTPRGNVLASHSPMSPESIKMTHRHSPMNLMGTGRHSSSSLSSHASSE AGNMVMLGDGSMGDAPEDLYHHMQLAYPNPRYQGSVTNVSVLSSSQASPSSSSLSSTHSAPSQMITSAPSSARGSPSLPDKYRHAREMML LLPTYRDRPSSAMYPAAILENGQPPNFQRALFQQVVGACKPCSDPNLSVAEKGHYSLHFDAFHHPLGDTPPALPARTLRKSPLHPIPASP TSPQSGLDGSNSTLSGSASSGVSSLSESNFGHSSEAPPRTDTMDSMPSQAWNADEDLEPPYLPVHYSLSESAVLDSIKAQPCRSHSAPGC -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:50655215/chr3:50927457) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAPKAPK3 | DOCK3 |
| FUNCTION: Stress-activated serine/threonine-protein kinase involved in cytokines production, endocytosis, cell migration, chromatin remodeling and transcriptional regulation. Following stress, it is phosphorylated and activated by MAP kinase p38-alpha/MAPK14, leading to phosphorylation of substrates. Phosphorylates serine in the peptide sequence, Hyd-X-R-X(2)-S, where Hyd is a large hydrophobic residue. MAPKAPK2 and MAPKAPK3, share the same function and substrate specificity, but MAPKAPK3 kinase activity and level in protein expression are lower compared to MAPKAPK2. Phosphorylates HSP27/HSPB1, KRT18, KRT20, RCSD1, RPS6KA3, TAB3 and TTP/ZFP36. Mediates phosphorylation of HSP27/HSPB1 in response to stress, leading to dissociate HSP27/HSPB1 from large small heat-shock protein (sHsps) oligomers and impair their chaperone activities and ability to protect against oxidative stress effectively. Involved in inflammatory response by regulating tumor necrosis factor (TNF) and IL6 production post-transcriptionally: acts by phosphorylating AU-rich elements (AREs)-binding proteins, such as TTP/ZFP36, leading to regulate the stability and translation of TNF and IL6 mRNAs. Phosphorylation of TTP/ZFP36, a major post-transcriptional regulator of TNF, promotes its binding to 14-3-3 proteins and reduces its ARE mRNA affinity leading to inhibition of dependent degradation of ARE-containing transcript. Involved in toll-like receptor signaling pathway (TLR) in dendritic cells: required for acute TLR-induced macropinocytosis by phosphorylating and activating RPS6KA3. Also acts as a modulator of Polycomb-mediated repression. {ECO:0000269|PubMed:10383393, ECO:0000269|PubMed:15563468, ECO:0000269|PubMed:18021073, ECO:0000269|PubMed:20599781, ECO:0000269|PubMed:8626550, ECO:0000269|PubMed:8774846}. | FUNCTION: Potential guanine nucleotide exchange factor (GEF). GEF proteins activate some small GTPases by exchanging bound GDP for free GTP. Its interaction with presenilin proteins as well as its ability to stimulate Tau/MAPT phosphorylation suggest that it may be involved in Alzheimer disease. Ectopic expression in nerve cells decreases the secretion of amyloid-beta APBA1 protein and lowers the rate of cell-substratum adhesion, suggesting that it may affect the function of some small GTPase involved in the regulation of actin cytoskeleton or cell adhesion receptors (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 50_58 | 73.0 | 383.0 | Nucleotide binding | ATP |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 50_58 | 73.0 | 383.0 | Nucleotide binding | ATP |
| Tgene | DOCK3 | chr3:50655215 | chr3:50927457 | ENST00000266037 | 2 | 53 | 1731_1768 | 54.0 | 2031.0 | Compositional bias | Note=Ser-rich | |
| Tgene | DOCK3 | chr3:50655215 | chr3:50927457 | ENST00000266037 | 2 | 53 | 1228_1635 | 54.0 | 2031.0 | Domain | DOCKER | |
| Tgene | DOCK3 | chr3:50655215 | chr3:50927457 | ENST00000266037 | 2 | 53 | 421_599 | 54.0 | 2031.0 | Domain | C2 DOCK-type | |
| Tgene | DOCK3 | chr3:50655215 | chr3:50927457 | ENST00000266037 | 2 | 53 | 1970_1976 | 54.0 | 2031.0 | Motif | SH3-binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 44_304 | 73.0 | 383.0 | Domain | Protein kinase |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 44_304 | 73.0 | 383.0 | Domain | Protein kinase |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 335_344 | 73.0 | 383.0 | Motif | Nuclear export signal (NES) |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 350_353 | 73.0 | 383.0 | Motif | Bipartite nuclear localization signal 1 |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 364_368 | 73.0 | 383.0 | Motif | Bipartite nuclear localization signal 2 |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 335_344 | 73.0 | 383.0 | Motif | Nuclear export signal (NES) |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 350_353 | 73.0 | 383.0 | Motif | Bipartite nuclear localization signal 1 |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 364_368 | 73.0 | 383.0 | Motif | Bipartite nuclear localization signal 2 |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 307_343 | 73.0 | 383.0 | Region | Autoinhibitory helix |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000357955 | + | 2 | 11 | 345_369 | 73.0 | 383.0 | Region | p38 MAPK-binding site |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 307_343 | 73.0 | 383.0 | Region | Autoinhibitory helix |
| Hgene | MAPKAPK3 | chr3:50655215 | chr3:50927457 | ENST00000446044 | + | 4 | 13 | 345_369 | 73.0 | 383.0 | Region | p38 MAPK-binding site |
| Tgene | DOCK3 | chr3:50655215 | chr3:50927457 | ENST00000266037 | 2 | 53 | 6_67 | 54.0 | 2031.0 | Domain | SH3 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MAPKAPK3 | |
| DOCK3 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MAPKAPK3-DOCK3 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MAPKAPK3-DOCK3 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

