| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MCM5-LYN |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MCM5-LYN | FusionPDB ID: 52233 | FusionGDB2.0 ID: 52233 | Hgene | Tgene | Gene symbol | MCM5 | LYN | Gene ID | 4174 | 4067 |
| Gene name | minichromosome maintenance complex component 5 | LYN proto-oncogene, Src family tyrosine kinase | |
| Synonyms | CDC46|MGORS8|P1-CDC46 | JTK8|p53Lyn|p56Lyn | |
| Cytomap | 22q12.3 | 8q12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | DNA replication licensing factor MCM5CDC46 homologMCM5 minichromosome maintenance deficient 5, cell division cycle 46minichromosome maintenance deficient 5 (cell division cycle 46) | tyrosine-protein kinase Lynlck/Yes-related novel protein tyrosine kinasev-yes-1 Yamaguchi sarcoma viral related oncogene homolog | |
| Modification date | 20200315 | 20200327 | |
| UniProtAcc | P33992 | P0DP58 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000216122, ENST00000382011, ENST00000465557, | ENST00000420292, ENST00000519728, ENST00000520220, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 17 X 12 X 7=1428 | 13 X 14 X 7=1274 |
| # samples | 22 | 15 | |
| ** MAII score | log2(22/1428*10)=-2.69842055050444 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1274*10)=-3.08633087176042 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MCM5 [Title/Abstract] AND LYN [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MCM5(35799535)-LYN(56910905), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | MCM5-LYN seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MCM5-LYN seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | LYN | GO:0006468 | protein phosphorylation | 11517336 |
| Tgene | LYN | GO:0006974 | cellular response to DNA damage stimulus | 10891478|11517336 |
| Tgene | LYN | GO:0018108 | peptidyl-tyrosine phosphorylation | 7682714|11782428 |
| Tgene | LYN | GO:0046777 | protein autophosphorylation | 7682714 |
| Tgene | LYN | GO:0051272 | positive regulation of cellular component movement | 16467205 |
| Tgene | LYN | GO:0070304 | positive regulation of stress-activated protein kinase signaling cascade | 10891478 |
 Fusion gene breakpoints across MCM5 (5'-gene) Fusion gene breakpoints across MCM5 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
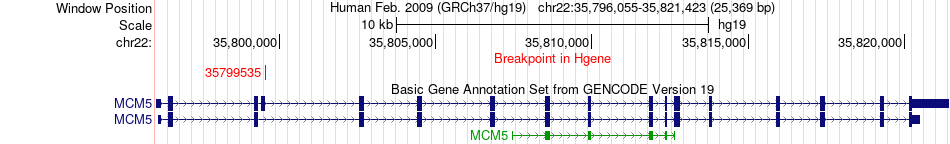 |
 Fusion gene breakpoints across LYN (3'-gene) Fusion gene breakpoints across LYN (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
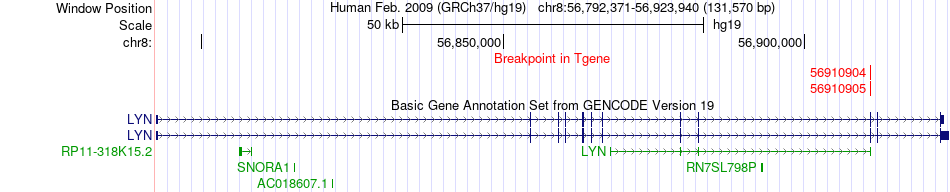 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-25-1329-01A | MCM5 | chr22 | 35799535 | - | LYN | chr8 | 56910905 | + |
| ChimerDB4 | OV | TCGA-25-1329-01A | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910905 | + |
| ChimerDB4 | OV | TCGA-25-1329 | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910904 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000216122 | MCM5 | chr22 | 35799535 | + | ENST00000519728 | LYN | chr8 | 56910905 | + | 1528 | 577 | 109 | 1065 | 318 |
| ENST00000216122 | MCM5 | chr22 | 35799535 | + | ENST00000520220 | LYN | chr8 | 56910905 | + | 2337 | 577 | 109 | 1065 | 318 |
| ENST00000216122 | MCM5 | chr22 | 35799535 | + | ENST00000519728 | LYN | chr8 | 56910904 | + | 1528 | 577 | 109 | 1065 | 318 |
| ENST00000216122 | MCM5 | chr22 | 35799535 | + | ENST00000520220 | LYN | chr8 | 56910904 | + | 2337 | 577 | 109 | 1065 | 318 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000216122 | ENST00000519728 | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910905 | + | 0.003773092 | 0.9962269 |
| ENST00000216122 | ENST00000520220 | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910905 | + | 0.002383934 | 0.99761605 |
| ENST00000216122 | ENST00000519728 | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910904 | + | 0.003773092 | 0.9962269 |
| ENST00000216122 | ENST00000520220 | MCM5 | chr22 | 35799535 | + | LYN | chr8 | 56910904 | + | 0.002383934 | 0.99761605 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >52233_52233_1_MCM5-LYN_MCM5_chr22_35799535_ENST00000216122_LYN_chr8_56910904_ENST00000519728_length(amino acids)=318AA_BP=156 MSPLVCEVRKTRGAVMSGFDDPGIFYSDSFGGDAQADEGQARKSQLQRRFKEFLRQYRVGTDRTGFTFKYRDELKRHYNLGEYWIEVEME DLASFDEDLADYLYKQPAEHLQLLEEAAKEVADEVTRPRPSGEEVLQDIQVMLKSDASPSSIRSLKIAEGMAYIERKNYIHRDLRAANVL VSESLMCKIADFGLARVIEDNEYTAREGAKFPIKWTAPEAINFGCFTIKSDVWSFGILLYEIVTYGKIPYPGRTNADVMTALSQGYRMPR -------------------------------------------------------------- >52233_52233_2_MCM5-LYN_MCM5_chr22_35799535_ENST00000216122_LYN_chr8_56910904_ENST00000520220_length(amino acids)=318AA_BP=156 MSPLVCEVRKTRGAVMSGFDDPGIFYSDSFGGDAQADEGQARKSQLQRRFKEFLRQYRVGTDRTGFTFKYRDELKRHYNLGEYWIEVEME DLASFDEDLADYLYKQPAEHLQLLEEAAKEVADEVTRPRPSGEEVLQDIQVMLKSDASPSSIRSLKIAEGMAYIERKNYIHRDLRAANVL VSESLMCKIADFGLARVIEDNEYTAREGAKFPIKWTAPEAINFGCFTIKSDVWSFGILLYEIVTYGKIPYPGRTNADVMTALSQGYRMPR -------------------------------------------------------------- >52233_52233_3_MCM5-LYN_MCM5_chr22_35799535_ENST00000216122_LYN_chr8_56910905_ENST00000519728_length(amino acids)=318AA_BP=156 MSPLVCEVRKTRGAVMSGFDDPGIFYSDSFGGDAQADEGQARKSQLQRRFKEFLRQYRVGTDRTGFTFKYRDELKRHYNLGEYWIEVEME DLASFDEDLADYLYKQPAEHLQLLEEAAKEVADEVTRPRPSGEEVLQDIQVMLKSDASPSSIRSLKIAEGMAYIERKNYIHRDLRAANVL VSESLMCKIADFGLARVIEDNEYTAREGAKFPIKWTAPEAINFGCFTIKSDVWSFGILLYEIVTYGKIPYPGRTNADVMTALSQGYRMPR -------------------------------------------------------------- >52233_52233_4_MCM5-LYN_MCM5_chr22_35799535_ENST00000216122_LYN_chr8_56910905_ENST00000520220_length(amino acids)=318AA_BP=156 MSPLVCEVRKTRGAVMSGFDDPGIFYSDSFGGDAQADEGQARKSQLQRRFKEFLRQYRVGTDRTGFTFKYRDELKRHYNLGEYWIEVEME DLASFDEDLADYLYKQPAEHLQLLEEAAKEVADEVTRPRPSGEEVLQDIQVMLKSDASPSSIRSLKIAEGMAYIERKNYIHRDLRAANVL VSESLMCKIADFGLARVIEDNEYTAREGAKFPIKWTAPEAINFGCFTIKSDVWSFGILLYEIVTYGKIPYPGRTNADVMTALSQGYRMPR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:35799535/chr8:56910905) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MCM5 | LYN |
| FUNCTION: Acts as component of the MCM2-7 complex (MCM complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the MCM2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. {ECO:0000269|PubMed:16899510}. | FUNCTION: Acts in different tissues through interaction to nicotinic acetylcholine receptors (nAChRs) (PubMed:21252236). The proposed role as modulator of nAChR activity seems to be dependent on the nAChR subtype and stoichiometry, and to involve an effect on nAChR trafficking and its cell surface expression, and on single channel properties of the nAChR inserted in the plasma membrane. Modulates functional properties of nicotinic acetylcholine receptors (nAChRs) to prevent excessive excitation, and hence neurodegeneration. Enhances desensitization by increasing both the rate and extent of desensitization of alpha-4:beta-2-containing nAChRs and slowing recovery from desensitization. Promotes large amplitude ACh-evoked currents through alpha-4:beta-2 nAChRs. Is involved in regulation of the nAChR pentameric assembly in the endoplasmic reticulum. Shifts stoichiometry from high sensitivity alpha-4(2):beta-2(3) to low sensitivity alpha-4(3):beta-2(2) nAChR (By similarity). In vitro modulates alpha-3:beta-4-containing nAChRs. Reduces cell surface expression of (alpha-3:beta-4)(2):beta-4 and (alpha-3:beta-4)(2):alpha-5 nAChRs suggesting an interaction with nAChR alpha-3(-):(+)beta-4 subunit interfaces and an allosteric mode. Corresponding single channel effects characterized by decreased unitary conductance, altered burst proportions and enhanced desensitization/inactivation seem to depend on nAChR alpha:alpha subunit interfaces and are greater in (alpha-3:beta-2)(2):alpha-3 when compared to (alpha-3:beta-2)(2):alpha-5 nAChRs (PubMed:28100642). Prevents plasticity in the primary visual cortex late in life (By similarity). {ECO:0000250|UniProtKB:P0DP60, ECO:0000269|PubMed:21252236, ECO:0000269|PubMed:28100642}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MCM5 | chr22:35799535 | chr8:56910904 | ENST00000216122 | + | 4 | 17 | 331_537 | 141.0 | 735.0 | Domain | Note=MCM |
| Hgene | MCM5 | chr22:35799535 | chr8:56910905 | ENST00000216122 | + | 4 | 17 | 331_537 | 141.0 | 735.0 | Domain | Note=MCM |
| Hgene | MCM5 | chr22:35799535 | chr8:56910904 | ENST00000216122 | + | 4 | 17 | 381_388 | 141.0 | 735.0 | Nucleotide binding | ATP |
| Hgene | MCM5 | chr22:35799535 | chr8:56910905 | ENST00000216122 | + | 4 | 17 | 381_388 | 141.0 | 735.0 | Nucleotide binding | ATP |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000519728 | 9 | 13 | 129_226 | 350.0 | 513.0 | Domain | SH2 | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000519728 | 9 | 13 | 247_501 | 350.0 | 513.0 | Domain | Protein kinase | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000519728 | 9 | 13 | 63_123 | 350.0 | 513.0 | Domain | SH3 | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000520220 | 9 | 13 | 129_226 | 329.0 | 492.0 | Domain | SH2 | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000520220 | 9 | 13 | 247_501 | 329.0 | 492.0 | Domain | Protein kinase | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000520220 | 9 | 13 | 63_123 | 329.0 | 492.0 | Domain | SH3 | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000519728 | 9 | 13 | 129_226 | 350.0 | 513.0 | Domain | SH2 | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000519728 | 9 | 13 | 247_501 | 350.0 | 513.0 | Domain | Protein kinase | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000519728 | 9 | 13 | 63_123 | 350.0 | 513.0 | Domain | SH3 | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000520220 | 9 | 13 | 129_226 | 329.0 | 492.0 | Domain | SH2 | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000520220 | 9 | 13 | 247_501 | 329.0 | 492.0 | Domain | Protein kinase | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000520220 | 9 | 13 | 63_123 | 329.0 | 492.0 | Domain | SH3 | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000519728 | 9 | 13 | 253_261 | 350.0 | 513.0 | Nucleotide binding | ATP | |
| Tgene | LYN | chr22:35799535 | chr8:56910904 | ENST00000520220 | 9 | 13 | 253_261 | 329.0 | 492.0 | Nucleotide binding | ATP | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000519728 | 9 | 13 | 253_261 | 350.0 | 513.0 | Nucleotide binding | ATP | |
| Tgene | LYN | chr22:35799535 | chr8:56910905 | ENST00000520220 | 9 | 13 | 253_261 | 329.0 | 492.0 | Nucleotide binding | ATP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MCM5 | |
| LYN |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MCM5-LYN |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MCM5-LYN |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

