| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MYH11-MYLK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MYH11-MYLK | FusionPDB ID: 56300 | FusionGDB2.0 ID: 56300 | Hgene | Tgene | Gene symbol | MYH11 | MYLK | Gene ID | 4629 | 4638 |
| Gene name | myosin heavy chain 11 | myosin light chain kinase | |
| Synonyms | AAT4|FAA4|SMHC|SMMHC | AAT7|KRP|MLCK|MLCK1|MLCK108|MLCK210|MMIHS|MSTP083|MYLK1|smMLCK | |
| Cytomap | 16p13.11 | 3q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | myosin-11epididymis secretory sperm binding proteinmyosin heavy chain, smooth muscle isoformmyosin, heavy chain 11, smooth musclemyosin, heavy polypeptide 11, smooth muscle | myosin light chain kinase, smooth musclekinase-related proteinmyosin, light polypeptide kinasesmooth muscle myosin light chain kinasetelokin | |
| Modification date | 20200322 | 20200320 | |
| UniProtAcc | P35749 | Q86YV6 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000300036, ENST00000396324, ENST00000452625, ENST00000576790, ENST00000573908, | ENST00000346322, ENST00000354792, ENST00000418370, ENST00000475616, ENST00000510775, ENST00000578202, ENST00000583087, ENST00000359169, ENST00000360304, ENST00000360772, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 31 X 31 X 8=7688 | 14 X 15 X 4=840 |
| # samples | 32 | 15 | |
| ** MAII score | log2(32/7688*10)=-4.58646452588639 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/840*10)=-2.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MYH11 [Title/Abstract] AND MYLK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MYH11(15809021)-MYLK(123383105), # samples:1 MYLK(123333009)-MYH11(15797744), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MYH11-MYLK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYH11-MYLK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYH11-MYLK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYH11-MYLK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYH11-MYLK seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. MYH11-MYLK seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. MYH11-MYLK seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MYLK | GO:0030335 | positive regulation of cell migration | 19826488 |
| Tgene | MYLK | GO:0051928 | positive regulation of calcium ion transport | 16284075 |
| Tgene | MYLK | GO:0071476 | cellular hypotonic response | 11976941 |
| Tgene | MYLK | GO:0090303 | positive regulation of wound healing | 15825080 |
 Fusion gene breakpoints across MYH11 (5'-gene) Fusion gene breakpoints across MYH11 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
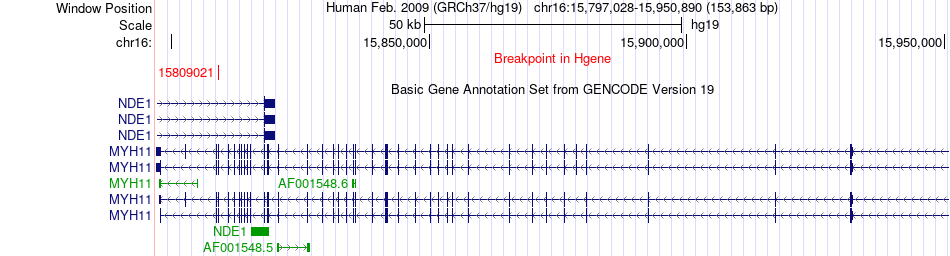 |
 Fusion gene breakpoints across MYLK (3'-gene) Fusion gene breakpoints across MYLK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
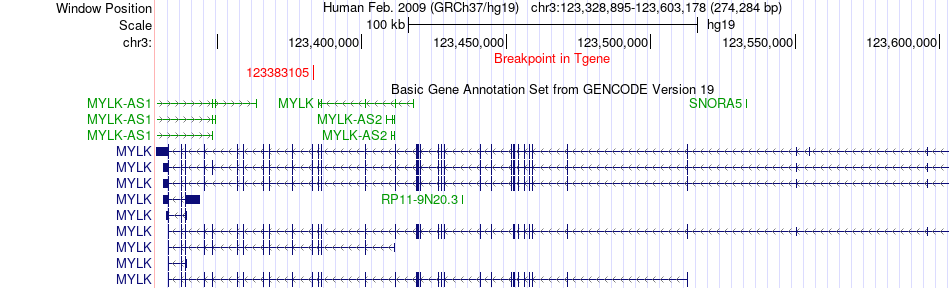 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-3B-A9HR-01A | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000452625 | MYH11 | chr16 | 15809021 | - | ENST00000360772 | MYLK | chr3 | 123383105 | - | 11539 | 5722 | 61 | 7482 | 2473 |
| ENST00000452625 | MYH11 | chr16 | 15809021 | - | ENST00000359169 | MYLK | chr3 | 123383105 | - | 9290 | 5722 | 61 | 7482 | 2473 |
| ENST00000452625 | MYH11 | chr16 | 15809021 | - | ENST00000360304 | MYLK | chr3 | 123383105 | - | 9443 | 5722 | 61 | 7635 | 2524 |
| ENST00000576790 | MYH11 | chr16 | 15809021 | - | ENST00000360772 | MYLK | chr3 | 123383105 | - | 11540 | 5723 | 83 | 7483 | 2466 |
| ENST00000576790 | MYH11 | chr16 | 15809021 | - | ENST00000359169 | MYLK | chr3 | 123383105 | - | 9291 | 5723 | 83 | 7483 | 2466 |
| ENST00000576790 | MYH11 | chr16 | 15809021 | - | ENST00000360304 | MYLK | chr3 | 123383105 | - | 9444 | 5723 | 83 | 7636 | 2517 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000452625 | ENST00000360772 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.00446648 | 0.99553347 |
| ENST00000452625 | ENST00000359169 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.009958294 | 0.99004173 |
| ENST00000452625 | ENST00000360304 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.008010932 | 0.9919891 |
| ENST00000576790 | ENST00000360772 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.004579956 | 0.9954201 |
| ENST00000576790 | ENST00000359169 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.010204897 | 0.98979515 |
| ENST00000576790 | ENST00000360304 | MYH11 | chr16 | 15809021 | - | MYLK | chr3 | 123383105 | - | 0.008210028 | 0.99178994 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >56300_56300_1_MYH11-MYLK_MYH11_chr16_15809021_ENST00000452625_MYLK_chr3_123383105_ENST00000359169_length(amino acids)=2473AA_BP=1887 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITQGPSFAYGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTG YIVGANIETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQL SILKVVSSVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFR WILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIER PNNPPGVLALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKF VADLWKDVDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGV LEGIRICRQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQ AMCRGYLARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQ LTEEKNLLQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKV TAEAKIKKLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDA SDFHEQIADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTE LEDTLDSTATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLG QAKQEVEHKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNV STKLRQLEEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNR LQQELDDLVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEME DLVSSKDDVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETE LEDERKQRALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQE DLAAAERARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNE SARQQLERQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQE SEHMKVENSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEI WDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKV SDFYDIEERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERI IDEDFELTERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGY ATDMWSIGVICYILNRLDCTQCLQHPWLMKDTKNMEAKKLSKDRMKKYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPL NAEKLESEEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEVVEGSAARFDCKIEGYPDPEVVWFKDDQSIRESRHFQIDYDEDGNCSLIISD -------------------------------------------------------------- >56300_56300_2_MYH11-MYLK_MYH11_chr16_15809021_ENST00000452625_MYLK_chr3_123383105_ENST00000360304_length(amino acids)=2524AA_BP=1887 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITQGPSFAYGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTG YIVGANIETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQL SILKVVSSVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFR WILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIER PNNPPGVLALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKF VADLWKDVDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGV LEGIRICRQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQ AMCRGYLARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQ LTEEKNLLQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKV TAEAKIKKLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDA SDFHEQIADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTE LEDTLDSTATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLG QAKQEVEHKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNV STKLRQLEEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNR LQQELDDLVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEME DLVSSKDDVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETE LEDERKQRALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQE DLAAAERARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNE SARQQLERQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQE SEHMKVENSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEI WDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKV SDFYDIEERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERI IDEDFELTERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGY ATDMWSIGVICYILVSGLSPFMGDNDNETLANVTSATWDFDDEAFDEISDDAKDFISNLLKKDMKNRLDCTQCLQHPWLMKDTKNMEAKK LSKDRMKKYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPLNAEKLESEEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEV VEGSAARFDCKIEGYPDPEVVWFKDDQSIRESRHFQIDYDEDGNCSLIISDVCGDDDAKYTCKAVNSLGEATCTAELIVETMEEGEGEGE -------------------------------------------------------------- >56300_56300_3_MYH11-MYLK_MYH11_chr16_15809021_ENST00000452625_MYLK_chr3_123383105_ENST00000360772_length(amino acids)=2473AA_BP=1887 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITQGPSFAYGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTG YIVGANIETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQL SILKVVSSVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFR WILTRVNKALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIER PNNPPGVLALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKF VADLWKDVDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGV LEGIRICRQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQ AMCRGYLARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQ LTEEKNLLQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKV TAEAKIKKLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDA SDFHEQIADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTE LEDTLDSTATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLG QAKQEVEHKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNV STKLRQLEEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNR LQQELDDLVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEME DLVSSKDDVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETE LEDERKQRALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQE DLAAAERARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNE SARQQLERQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQE SEHMKVENSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEI WDSANKTWKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKV SDFYDIEERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERI IDEDFELTERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGY ATDMWSIGVICYILNRLDCTQCLQHPWLMKDTKNMEAKKLSKDRMKKYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPL NAEKLESEEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEVVEGSAARFDCKIEGYPDPEVVWFKDDQSIRESRHFQIDYDEDGNCSLIISD -------------------------------------------------------------- >56300_56300_4_MYH11-MYLK_MYH11_chr16_15809021_ENST00000576790_MYLK_chr3_123383105_ENST00000359169_length(amino acids)=2466AA_BP=1880 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTGYIVGANI ETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQLSILKVVS SVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFRWILTRVN KALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIERPNNPPGV LALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKFVADLWKD VDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGVLEGIRIC RQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQAMCRGYL ARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQLTEEKNL LQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIK KLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQI ADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTELEDTLDS TATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLGQAKQEVE HKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNVSTKLRQL EEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNRLQQELDD LVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEMEDLVSSKD DVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETELEDERKQ RALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQEDLAAAER ARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNESARQQLE RQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQESEHMKVE NSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKT WKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKVSDFYDIE ERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERIIDEDFEL TERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGYATDMWSI GVICYILNRLDCTQCLQHPWLMKDTKNMEAKKLSKDRMKKYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPLNAEKLES EEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEVVEGSAARFDCKIEGYPDPEVVWFKDDQSIRESRHFQIDYDEDGNCSLIISDVCGDDDA -------------------------------------------------------------- >56300_56300_5_MYH11-MYLK_MYH11_chr16_15809021_ENST00000576790_MYLK_chr3_123383105_ENST00000360304_length(amino acids)=2517AA_BP=1880 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTGYIVGANI ETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQLSILKVVS SVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFRWILTRVN KALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIERPNNPPGV LALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKFVADLWKD VDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGVLEGIRIC RQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQAMCRGYL ARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQLTEEKNL LQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIK KLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQI ADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTELEDTLDS TATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLGQAKQEVE HKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNVSTKLRQL EEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNRLQQELDD LVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEMEDLVSSKD DVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETELEDERKQ RALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQEDLAAAER ARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNESARQQLE RQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQESEHMKVE NSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKT WKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKVSDFYDIE ERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERIIDEDFEL TERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGYATDMWSI GVICYILVSGLSPFMGDNDNETLANVTSATWDFDDEAFDEISDDAKDFISNLLKKDMKNRLDCTQCLQHPWLMKDTKNMEAKKLSKDRMK KYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPLNAEKLESEEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEVVEGSAAR -------------------------------------------------------------- >56300_56300_6_MYH11-MYLK_MYH11_chr16_15809021_ENST00000576790_MYLK_chr3_123383105_ENST00000360772_length(amino acids)=2466AA_BP=1880 MEIWDQQGTMAQKGQLSDDEKFLFVDKNFINSPVAQADWAAKRLVWVPSEKQGFEAASIKEEKGDEVVVELVENGKKVTVGKDDIQKMNP PKFSKVEDMAELTCLNEASVLHNLRERYFSGLIYTYSGLFCVVVNPYKHLPIYSEKIVDMYKGKKRHEMPPHIYAIADTAYRSMLQDRED QSILCTGESGAGKTENTKKVIQYLAVVASSHKGKKDTSITGELEKQLLQANPILEAFGNAKTVKNDNSSRFGKFIRINFDVTGYIVGANI ETYLLEKSRAIRQARDERTFHIFYYMIAGAKEKMRSDLLLEGFNNYTFLSNGFVPIPAAQDDEMFQETVEAMAIMGFSEEEQLSILKVVS SVLQLGNIVFKKERNTDQASMPDNTAAQKVCHLMGINVTDFTRSILTPRIKVGRDVVQKAQTKEQADFAVEALAKATYERLFRWILTRVN KALDKTHRQGASFLGILDIAGFEIFEVNSFEQLCINYTNEKLQQLFNHTMFILEQEEYQREGIEWNFIDFGLDLQPCIELIERPNNPPGV LALLDEECWFPKATDKSFVEKLCTEQGSHPKFQKPKQLKDKTEFSIIHYAGKVDYNASAWLTKNMDPLNDNVTSLLNASSDKFVADLWKD VDRIVGLDQMAKMTESSLPSASKTKKGMFRTVGQLYKEQLGKLMTTLRNTTPNFVRCIIPNHEKRSGKLDAFLVLEQLRCNGVLEGIRIC RQGFPNRIVFQEFRQRYEILAANAIPKGFMDGKQACILMIKALELDPNLYRIGQSKIFFRTGVLAHLEEERDLKITDVIMAFQAMCRGYL ARKAFAKRQQQLTAMKVIQRNCAAYLKLRNWQWWRLFTKVKPLLQVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQLTEEKNL LQEQLQAETELYAEAEEMRVRLAAKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIK KLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQI ADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLGEELEALKTELEDTLDS TATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLGQAKQEVE HKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLSSQLQDTQELLQEETRQKLNVSTKLRQL EEERNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENLTQQYEEKAAAYDKLEKTKNRLQQELDD LVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNISSKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEMEDLVSSKD DVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETELEDERKQ RALAAAAKKKLEGDLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKSLEADLMQLQEDLAAAER ARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNESARQQLE RQNKELRSKLHEMEGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKMAEQYKEQIQESEHMKVE NSENGSKLTILAARQEHCGCYTLLVENKLGSRQAQVNLTVVDKPDPPAGTPCASDIRSSSLTLSWYGSSYDGGSAVQSYSIEIWDSANKT WKELATCRSTSFNVQDLLPDHEYKFRVRAINVYGTSEPSQESELTTVGEKPEEPKDEVEVSDDDEKEPEVDYRTVTINTEQKVSDFYDIE ERLGSGKFGQVFRLVEKKTRKVWAGKFFKAYSAKEKENIRQEISIMNCLHHPKLVQCVDAFEEKANIVMVLEIVSGGELFERIIDEDFEL TERECIKYMRQISEGVEYIHKQGIVHLDLKPENIMCVNKTGTRIKLIDFGLARRLENAGSLKVLFGTPEFVAPEVINYEPIGYATDMWSI GVICYILNRLDCTQCLQHPWLMKDTKNMEAKKLSKDRMKKYMARRKWQKTGNAVRAIGRLSSMAMISGLSGRKSSTGSPTSPLNAEKLES EEDVSQAFLEAVAEEKPHVKPYFSKTIRDLEVVEGSAARFDCKIEGYPDPEVVWFKDDQSIRESRHFQIDYDEDGNCSLIISDVCGDDDA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:15809021/chr3:123383105) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MYH11 | MYLK |
| FUNCTION: Muscle contraction. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 31_81 | 1871.0 | 1973.0 | Domain | Myosin N-terminal SH3-like |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 786_815 | 1871.0 | 1973.0 | Domain | IQ |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 85_783 | 1871.0 | 1973.0 | Domain | Myosin motor |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 31_81 | 1878.0 | 1980.0 | Domain | Myosin N-terminal SH3-like |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 786_815 | 1878.0 | 1980.0 | Domain | IQ |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 85_783 | 1878.0 | 1980.0 | Domain | Myosin motor |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 31_81 | 1878.0 | 2132.0 | Domain | Myosin N-terminal SH3-like |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 786_815 | 1878.0 | 2132.0 | Domain | IQ |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 85_783 | 1878.0 | 2132.0 | Domain | Myosin motor |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 31_81 | 1871.0 | 1912.6666666666667 | Domain | Myosin N-terminal SH3-like |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 786_815 | 1871.0 | 1912.6666666666667 | Domain | IQ |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 85_783 | 1871.0 | 1912.6666666666667 | Domain | Myosin motor |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 178_185 | 1871.0 | 1973.0 | Nucleotide binding | ATP |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 178_185 | 1878.0 | 1980.0 | Nucleotide binding | ATP |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 178_185 | 1878.0 | 2132.0 | Nucleotide binding | ATP |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 178_185 | 1871.0 | 1912.6666666666667 | Nucleotide binding | ATP |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 661_683 | 1871.0 | 1973.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 762_776 | 1871.0 | 1973.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 661_683 | 1878.0 | 1980.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 762_776 | 1878.0 | 1980.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 661_683 | 1878.0 | 2132.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 762_776 | 1878.0 | 2132.0 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 661_683 | 1871.0 | 1912.6666666666667 | Region | Actin-binding |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 762_776 | 1871.0 | 1912.6666666666667 | Region | Actin-binding |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1906_1914 | 1208.0 | 1846.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1906_1914 | 1277.0 | 1864.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1906_1914 | 1277.0 | 1915.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1906_1914 | 1277.0 | 1864.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1906_1914 | 0 | 155.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1906_1914 | 1277.0 | 1915.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1906_1914 | 0 | 155.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1238_1326 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1334_1426 | 1208.0 | 1846.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1464_1719 | 1208.0 | 1846.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1809_1898 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1334_1426 | 1277.0 | 1864.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1464_1719 | 1277.0 | 1864.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1809_1898 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1334_1426 | 1277.0 | 1915.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1464_1719 | 1277.0 | 1915.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1809_1898 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1334_1426 | 1277.0 | 1864.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1464_1719 | 1277.0 | 1864.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1809_1898 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1098_1186 | 0 | 155.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1238_1326 | 0 | 155.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1334_1426 | 0 | 155.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1464_1719 | 0 | 155.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 161_249 | 0 | 155.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1809_1898 | 0 | 155.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 33_122 | 0 | 155.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 414_503 | 0 | 155.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 514_599 | 0 | 155.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 620_711 | 0 | 155.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 721_821 | 0 | 155.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1334_1426 | 1277.0 | 1915.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1464_1719 | 1277.0 | 1915.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1809_1898 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1098_1186 | 0 | 155.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1238_1326 | 0 | 155.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1334_1426 | 0 | 155.0 | Domain | Fibronectin type-III | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1464_1719 | 0 | 155.0 | Domain | Protein kinase | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 161_249 | 0 | 155.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1809_1898 | 0 | 155.0 | Domain | Note=Ig-like C2-type 9 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 33_122 | 0 | 155.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 414_503 | 0 | 155.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 514_599 | 0 | 155.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 620_711 | 0 | 155.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 721_821 | 0 | 155.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1470_1478 | 1208.0 | 1846.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1470_1478 | 1277.0 | 1864.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1470_1478 | 1277.0 | 1915.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1470_1478 | 1277.0 | 1864.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1470_1478 | 0 | 155.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1470_1478 | 1277.0 | 1915.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1470_1478 | 0 | 155.0 | Nucleotide binding | ATP | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1711_1774 | 1208.0 | 1846.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1711_1774 | 1277.0 | 1864.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1711_1774 | 1277.0 | 1915.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1711_1774 | 1277.0 | 1864.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1061_1460 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1711_1774 | 0 | 155.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 868_998 | 0 | 155.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 923_963 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 948_963 | 0 | 155.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 999_1063 | 0 | 155.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1711_1774 | 1277.0 | 1915.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1061_1460 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1711_1774 | 0 | 155.0 | Region | Note=Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 868_998 | 0 | 155.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 923_963 | 0 | 155.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 948_963 | 0 | 155.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 999_1063 | 0 | 155.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1004_1015 | 0 | 155.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1016_1027 | 0 | 155.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1028_1039 | 0 | 155.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1040_1051 | 0 | 155.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 1052_1063 | 0 | 155.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 868_895 | 0 | 155.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 896_923 | 0 | 155.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 924_951 | 0 | 155.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 952_979 | 0 | 155.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 980_998 | 0 | 155.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000418370 | 0 | 3 | 999_1003 | 0 | 155.0 | Repeat | Note=2-1%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1004_1015 | 0 | 155.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1016_1027 | 0 | 155.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1028_1039 | 0 | 155.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1040_1051 | 0 | 155.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 1052_1063 | 0 | 155.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 868_895 | 0 | 155.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 896_923 | 0 | 155.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 924_951 | 0 | 155.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 952_979 | 0 | 155.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 980_998 | 0 | 155.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000583087 | 0 | 4 | 999_1003 | 0 | 155.0 | Repeat | Note=2-1%3B truncated |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 844_1934 | 1871.0 | 1973.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 844_1934 | 1878.0 | 1980.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 844_1934 | 1878.0 | 2132.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 844_1934 | 1871.0 | 1912.6666666666667 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000300036 | - | 39 | 41 | 1935_1972 | 1871.0 | 1973.0 | Region | Note=C-terminal |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000396324 | - | 40 | 42 | 1935_1972 | 1878.0 | 1980.0 | Region | Note=C-terminal |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000452625 | - | 40 | 43 | 1935_1972 | 1878.0 | 2132.0 | Region | Note=C-terminal |
| Hgene | MYH11 | chr16:15809021 | chr3:123383105 | ENST00000576790 | - | 39 | 42 | 1935_1972 | 1871.0 | 1912.6666666666667 | Region | Note=C-terminal |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1098_1186 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 161_249 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 33_122 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 414_503 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 514_599 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 620_711 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 721_821 | 1208.0 | 1846.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1098_1186 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1238_1326 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 161_249 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 33_122 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 414_503 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 514_599 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 620_711 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 721_821 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1098_1186 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1238_1326 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 161_249 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 33_122 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 414_503 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 514_599 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 620_711 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 721_821 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1098_1186 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1238_1326 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 161_249 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 33_122 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 414_503 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 514_599 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 620_711 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 721_821 | 1277.0 | 1864.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1098_1186 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 7 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1238_1326 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 8 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 161_249 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 33_122 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 414_503 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 514_599 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 620_711 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 721_821 | 1277.0 | 1915.0 | Domain | Note=Ig-like C2-type 6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1061_1460 | 1208.0 | 1846.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 868_998 | 1208.0 | 1846.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 923_963 | 1208.0 | 1846.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 948_963 | 1208.0 | 1846.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 999_1063 | 1208.0 | 1846.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1061_1460 | 1277.0 | 1864.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 868_998 | 1277.0 | 1864.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 923_963 | 1277.0 | 1864.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 948_963 | 1277.0 | 1864.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 999_1063 | 1277.0 | 1864.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1061_1460 | 1277.0 | 1915.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 868_998 | 1277.0 | 1915.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 923_963 | 1277.0 | 1915.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 948_963 | 1277.0 | 1915.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 999_1063 | 1277.0 | 1915.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1061_1460 | 1277.0 | 1864.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 868_998 | 1277.0 | 1864.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 923_963 | 1277.0 | 1864.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 948_963 | 1277.0 | 1864.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 999_1063 | 1277.0 | 1864.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1061_1460 | 1277.0 | 1915.0 | Region | Actin-binding (calcium/calmodulin-insensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 868_998 | 1277.0 | 1915.0 | Region | Note=5 X 28 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 923_963 | 1277.0 | 1915.0 | Region | Actin-binding (calcium/calmodulin-sensitive) | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 948_963 | 1277.0 | 1915.0 | Region | Calmodulin-binding | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 999_1063 | 1277.0 | 1915.0 | Region | Note=6 X 12 AA approximate tandem repeats | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1004_1015 | 1208.0 | 1846.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1016_1027 | 1208.0 | 1846.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1028_1039 | 1208.0 | 1846.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1040_1051 | 1208.0 | 1846.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 1052_1063 | 1208.0 | 1846.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 868_895 | 1208.0 | 1846.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 896_923 | 1208.0 | 1846.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 924_951 | 1208.0 | 1846.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 952_979 | 1208.0 | 1846.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 980_998 | 1208.0 | 1846.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000346322 | 20 | 33 | 999_1003 | 1208.0 | 1846.0 | Repeat | Note=2-1%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1004_1015 | 1277.0 | 1864.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1016_1027 | 1277.0 | 1864.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1028_1039 | 1277.0 | 1864.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1040_1051 | 1277.0 | 1864.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 1052_1063 | 1277.0 | 1864.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 868_895 | 1277.0 | 1864.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 896_923 | 1277.0 | 1864.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 924_951 | 1277.0 | 1864.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 952_979 | 1277.0 | 1864.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 980_998 | 1277.0 | 1864.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000359169 | 21 | 33 | 999_1003 | 1277.0 | 1864.0 | Repeat | Note=2-1%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1004_1015 | 1277.0 | 1915.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1016_1027 | 1277.0 | 1915.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1028_1039 | 1277.0 | 1915.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1040_1051 | 1277.0 | 1915.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 1052_1063 | 1277.0 | 1915.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 868_895 | 1277.0 | 1915.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 896_923 | 1277.0 | 1915.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 924_951 | 1277.0 | 1915.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 952_979 | 1277.0 | 1915.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 980_998 | 1277.0 | 1915.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360304 | 21 | 34 | 999_1003 | 1277.0 | 1915.0 | Repeat | Note=2-1%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1004_1015 | 1277.0 | 1864.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1016_1027 | 1277.0 | 1864.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1028_1039 | 1277.0 | 1864.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1040_1051 | 1277.0 | 1864.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 1052_1063 | 1277.0 | 1864.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 868_895 | 1277.0 | 1864.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 896_923 | 1277.0 | 1864.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 924_951 | 1277.0 | 1864.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 952_979 | 1277.0 | 1864.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 980_998 | 1277.0 | 1864.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000360772 | 22 | 34 | 999_1003 | 1277.0 | 1864.0 | Repeat | Note=2-1%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1004_1015 | 1277.0 | 1915.0 | Repeat | Note=2-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1016_1027 | 1277.0 | 1915.0 | Repeat | Note=2-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1028_1039 | 1277.0 | 1915.0 | Repeat | Note=2-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1040_1051 | 1277.0 | 1915.0 | Repeat | Note=2-5 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 1052_1063 | 1277.0 | 1915.0 | Repeat | Note=2-6 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 868_895 | 1277.0 | 1915.0 | Repeat | Note=1-1 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 896_923 | 1277.0 | 1915.0 | Repeat | Note=1-2 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 924_951 | 1277.0 | 1915.0 | Repeat | Note=1-3 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 952_979 | 1277.0 | 1915.0 | Repeat | Note=1-4 | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 980_998 | 1277.0 | 1915.0 | Repeat | Note=1-5%3B truncated | |
| Tgene | MYLK | chr16:15809021 | chr3:123383105 | ENST00000475616 | 18 | 31 | 999_1003 | 1277.0 | 1915.0 | Repeat | Note=2-1%3B truncated |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MYH11 | 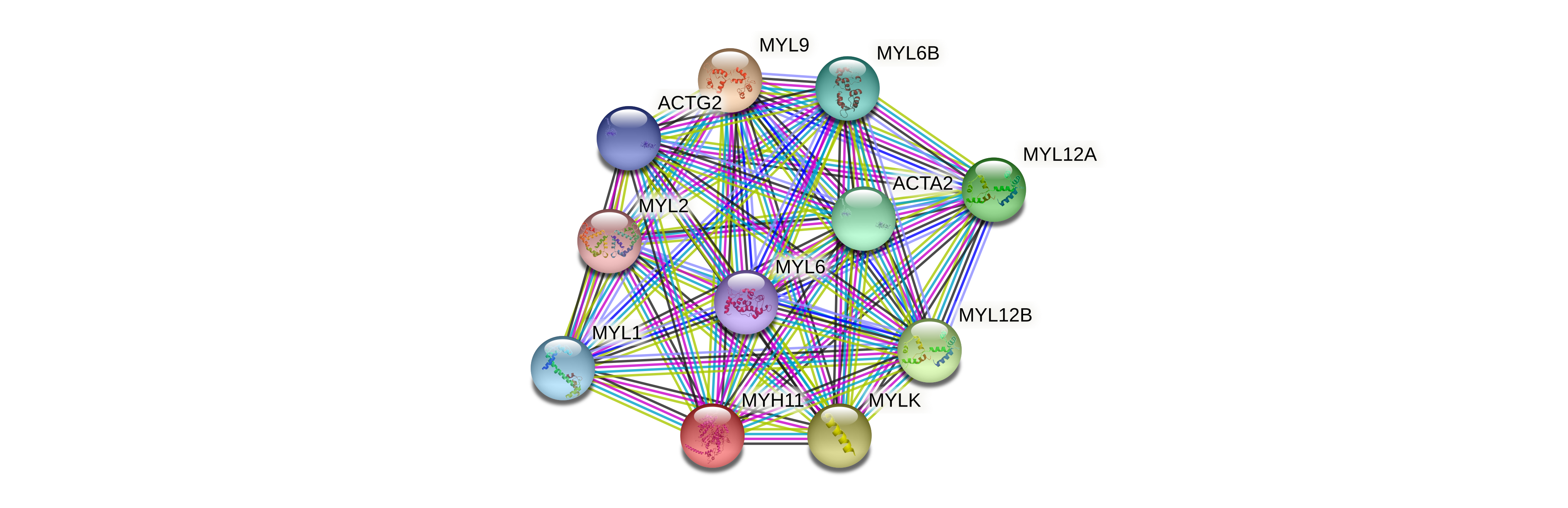 |
| MYLK |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MYH11-MYLK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MYH11-MYLK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | MYH11 | C4707243 | Familial thoracic aortic aneurysm and aortic dissection | 10 | CLINGEN;GENOMICS_ENGLAND |
| Hgene | MYH11 | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Hgene | MYH11 | C0023479 | Acute myelomonocytic leukemia | 2 | CTD_human;ORPHANET |
| Hgene | MYH11 | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Hgene | MYH11 | C1851504 | Aortic aneurysm, familial thoracic 4 | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | MYH11 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Hgene | MYH11 | C1608393 | Megacystis microcolon intestinal hypoperistalsis syndrome | 1 | GENOMICS_ENGLAND;ORPHANET |

