| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:APP-ABI1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: APP-ABI1 | FusionPDB ID: 5646 | FusionGDB2.0 ID: 5646 | Hgene | Tgene | Gene symbol | APP | ABI1 | Gene ID | 351 | 10006 |
| Gene name | amyloid beta precursor protein | abl interactor 1 | |
| Synonyms | AAA|ABETA|ABPP|AD1|APPI|CTFgamma|CVAP|PN-II|PN2|preA4 | ABI-1|ABLBP4|E3B1|NAP1BP|SSH3BP|SSH3BP1 | |
| Cytomap | 21q21.3 | 10p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | amyloid-beta precursor proteinalzheimer disease amyloid proteinamyloid beta (A4) precursor proteinamyloid beta A4 proteinamyloid precursor proteinbeta-amyloid peptidebeta-amyloid peptide(1-40)beta-amyloid peptide(1-42)beta-amyloid precursor protei | abl interactor 1Abelson interactor 1Abl-interactor protein 1 longabl-binding protein 4eps8 SH3 domain-binding proteininteractor protein AblBP4nap1 binding proteinspectrin SH3 domain-binding protein 1 | |
| Modification date | 20200329 | 20200327 | |
| UniProtAcc | Q8NEU8 | Q8IZP0 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000346798, ENST00000348990, ENST00000354192, ENST00000357903, ENST00000358918, ENST00000359726, ENST00000439274, ENST00000474136, ENST00000440126, ENST00000448388, | ENST00000473481, ENST00000346832, ENST00000376134, ENST00000376137, ENST00000376140, ENST00000490841, ENST00000536334, ENST00000355394, ENST00000359188, ENST00000376138, ENST00000376139, ENST00000376142, ENST00000376160, ENST00000376166, ENST00000376170, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 31 X 25 X 12=9300 | 14 X 8 X 5=560 |
| # samples | 33 | 13 | |
| ** MAII score | log2(33/9300*10)=-4.81669278663694 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/560*10)=-2.10691520391651 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: APP [Title/Abstract] AND ABI1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | APP(27542882)-ABI1(27066170), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | APP-ABI1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. APP-ABI1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. APP-ABI1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. APP-ABI1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | APP | GO:0001934 | positive regulation of protein phosphorylation | 11404397 |
| Hgene | APP | GO:0008285 | negative regulation of cell proliferation | 22944668 |
| Hgene | APP | GO:1905606 | regulation of presynapse assembly | 19726636 |
| Tgene | ABI1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 17101133 |
 Fusion gene breakpoints across APP (5'-gene) Fusion gene breakpoints across APP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
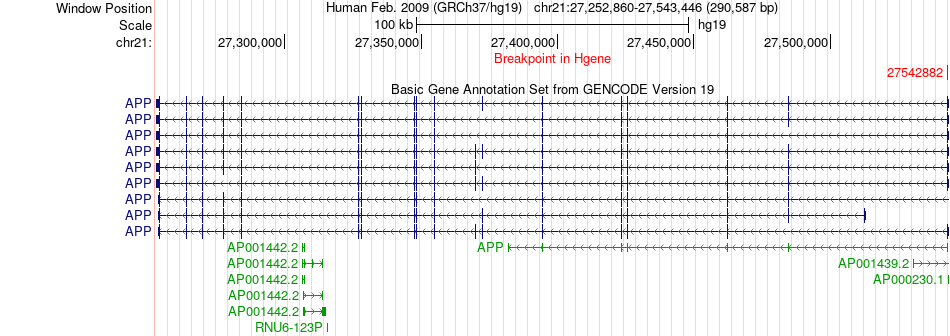 |
 Fusion gene breakpoints across ABI1 (3'-gene) Fusion gene breakpoints across ABI1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
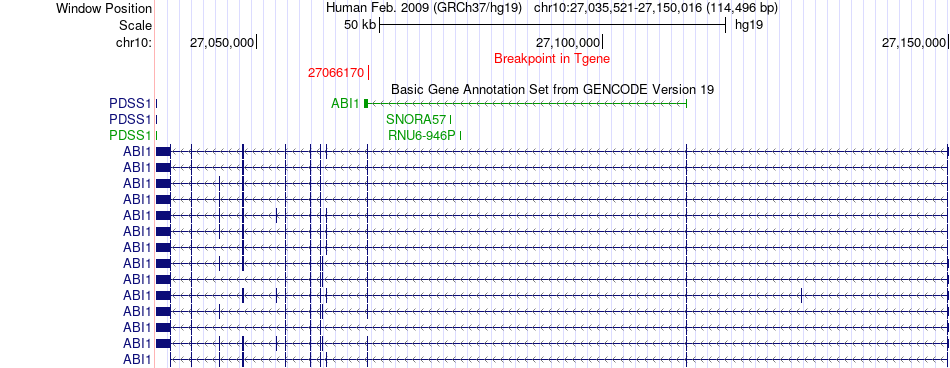 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-8525-01A | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3139 | 91 | 34 | 1161 | 375 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3226 | 91 | 34 | 1248 | 404 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3310 | 91 | 34 | 1332 | 432 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3214 | 91 | 34 | 1236 | 400 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3211 | 91 | 34 | 1233 | 399 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3124 | 91 | 34 | 1146 | 370 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3142 | 91 | 34 | 1164 | 376 |
| ENST00000346798 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3310 | 91 | 34 | 1335 | 433 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3232 | 184 | 127 | 1254 | 375 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3319 | 184 | 127 | 1341 | 404 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3403 | 184 | 127 | 1425 | 432 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3307 | 184 | 127 | 1329 | 400 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3304 | 184 | 127 | 1326 | 399 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3217 | 184 | 127 | 1239 | 370 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3235 | 184 | 127 | 1257 | 376 |
| ENST00000354192 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3403 | 184 | 127 | 1428 | 433 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3252 | 204 | 147 | 1274 | 375 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3339 | 204 | 147 | 1361 | 404 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3423 | 204 | 147 | 1445 | 432 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3327 | 204 | 147 | 1349 | 400 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3324 | 204 | 147 | 1346 | 399 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3237 | 204 | 147 | 1259 | 370 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3255 | 204 | 147 | 1277 | 376 |
| ENST00000348990 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3423 | 204 | 147 | 1448 | 433 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3272 | 224 | 167 | 1294 | 375 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3359 | 224 | 167 | 1381 | 404 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3443 | 224 | 167 | 1465 | 432 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3347 | 224 | 167 | 1369 | 400 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3344 | 224 | 167 | 1366 | 399 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3257 | 224 | 167 | 1279 | 370 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3275 | 224 | 167 | 1297 | 376 |
| ENST00000357903 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3443 | 224 | 167 | 1468 | 433 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3305 | 257 | 200 | 1327 | 375 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3392 | 257 | 200 | 1414 | 404 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3476 | 257 | 200 | 1498 | 432 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3380 | 257 | 200 | 1402 | 400 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3377 | 257 | 200 | 1399 | 399 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3290 | 257 | 200 | 1312 | 370 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3308 | 257 | 200 | 1330 | 376 |
| ENST00000358918 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3476 | 257 | 200 | 1501 | 433 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3305 | 257 | 200 | 1327 | 375 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3392 | 257 | 200 | 1414 | 404 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3476 | 257 | 200 | 1498 | 432 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3380 | 257 | 200 | 1402 | 400 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3377 | 257 | 200 | 1399 | 399 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3290 | 257 | 200 | 1312 | 370 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3308 | 257 | 200 | 1330 | 376 |
| ENST00000359726 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3476 | 257 | 200 | 1501 | 433 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376170 | ABI1 | chr10 | 27066170 | - | 3139 | 91 | 34 | 1161 | 375 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000359188 | ABI1 | chr10 | 27066170 | - | 3226 | 91 | 34 | 1248 | 404 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376142 | ABI1 | chr10 | 27066170 | - | 3310 | 91 | 34 | 1332 | 432 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376139 | ABI1 | chr10 | 27066170 | - | 3214 | 91 | 34 | 1236 | 400 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376160 | ABI1 | chr10 | 27066170 | - | 3211 | 91 | 34 | 1233 | 399 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376166 | ABI1 | chr10 | 27066170 | - | 3124 | 91 | 34 | 1146 | 370 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000376138 | ABI1 | chr10 | 27066170 | - | 3142 | 91 | 34 | 1164 | 376 |
| ENST00000439274 | APP | chr21 | 27542882 | - | ENST00000355394 | ABI1 | chr10 | 27066170 | - | 3310 | 91 | 34 | 1335 | 433 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000346798 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.52E-05 | 0.99996483 |
| ENST00000346798 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.20E-05 | 0.99995804 |
| ENST00000346798 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.10E-05 | 0.99993896 |
| ENST00000346798 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.19E-05 | 0.99995804 |
| ENST00000346798 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.16E-05 | 0.9999484 |
| ENST00000346798 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.42E-05 | 0.9999658 |
| ENST00000346798 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.06E-05 | 0.99996936 |
| ENST00000346798 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 9.02E-05 | 0.99990976 |
| ENST00000354192 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.84E-05 | 0.9999616 |
| ENST00000354192 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.57E-05 | 0.99995434 |
| ENST00000354192 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.77E-05 | 0.9999323 |
| ENST00000354192 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.62E-05 | 0.99995375 |
| ENST00000354192 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.65E-05 | 0.9999435 |
| ENST00000354192 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.76E-05 | 0.99996245 |
| ENST00000354192 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.34E-05 | 0.9999666 |
| ENST00000354192 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 0.000100837 | 0.99989915 |
| ENST00000348990 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.04E-05 | 0.9999596 |
| ENST00000348990 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.84E-05 | 0.9999516 |
| ENST00000348990 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 7.17E-05 | 0.99992824 |
| ENST00000348990 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.89E-05 | 0.9999511 |
| ENST00000348990 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.98E-05 | 0.99994016 |
| ENST00000348990 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.96E-05 | 0.9999604 |
| ENST00000348990 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.52E-05 | 0.99996483 |
| ENST00000348990 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 0.000106701 | 0.9998933 |
| ENST00000357903 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.09E-05 | 0.9999591 |
| ENST00000357903 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.89E-05 | 0.9999511 |
| ENST00000357903 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 7.23E-05 | 0.99992764 |
| ENST00000357903 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.93E-05 | 0.99995065 |
| ENST00000357903 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.05E-05 | 0.99993956 |
| ENST00000357903 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.01E-05 | 0.9999598 |
| ENST00000357903 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.56E-05 | 0.99996436 |
| ENST00000357903 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 0.000107478 | 0.9998925 |
| ENST00000358918 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.41E-05 | 0.9999559 |
| ENST00000358918 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.27E-05 | 0.9999473 |
| ENST00000358918 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 7.80E-05 | 0.99992204 |
| ENST00000358918 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.32E-05 | 0.99994683 |
| ENST00000358918 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.52E-05 | 0.9999348 |
| ENST00000358918 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.34E-05 | 0.9999566 |
| ENST00000358918 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.85E-05 | 0.9999615 |
| ENST00000358918 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 0.000115585 | 0.99988437 |
| ENST00000359726 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.41E-05 | 0.9999559 |
| ENST00000359726 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.27E-05 | 0.9999473 |
| ENST00000359726 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 7.80E-05 | 0.99992204 |
| ENST00000359726 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.32E-05 | 0.99994683 |
| ENST00000359726 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.52E-05 | 0.9999348 |
| ENST00000359726 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.34E-05 | 0.9999566 |
| ENST00000359726 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.85E-05 | 0.9999615 |
| ENST00000359726 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 0.000115585 | 0.99988437 |
| ENST00000439274 | ENST00000376170 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.52E-05 | 0.99996483 |
| ENST00000439274 | ENST00000359188 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.20E-05 | 0.99995804 |
| ENST00000439274 | ENST00000376142 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 6.10E-05 | 0.99993896 |
| ENST00000439274 | ENST00000376139 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 4.19E-05 | 0.99995804 |
| ENST00000439274 | ENST00000376160 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 5.16E-05 | 0.9999484 |
| ENST00000439274 | ENST00000376166 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.42E-05 | 0.9999658 |
| ENST00000439274 | ENST00000376138 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 3.06E-05 | 0.99996936 |
| ENST00000439274 | ENST00000355394 | APP | chr21 | 27542882 | - | ABI1 | chr10 | 27066170 | - | 9.02E-05 | 0.99990976 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >5646_5646_1_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_2_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_3_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_4_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_5_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_6_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_7_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_8_APP-ABI1_APP_chr21_27542882_ENST00000346798_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_9_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_10_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_11_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_12_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_13_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_14_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_15_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_16_APP-ABI1_APP_chr21_27542882_ENST00000348990_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_17_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_18_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_19_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_20_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_21_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_22_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_23_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_24_APP-ABI1_APP_chr21_27542882_ENST00000354192_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_25_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_26_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_27_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_28_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_29_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_30_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_31_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_32_APP-ABI1_APP_chr21_27542882_ENST00000357903_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_33_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_34_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_35_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_36_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_37_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_38_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_39_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_40_APP-ABI1_APP_chr21_27542882_ENST00000358918_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_41_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_42_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_43_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_44_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_45_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_46_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_47_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_48_APP-ABI1_APP_chr21_27542882_ENST00000359726_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- >5646_5646_49_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000355394_length(amino acids)=433AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKKWHKIIHGNNQP ARTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSS SIGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSA QPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPY -------------------------------------------------------------- >5646_5646_50_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000359188_length(amino acids)=404AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTP QIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDE -------------------------------------------------------------- >5646_5646_51_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376138_length(amino acids)=376AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPD DIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVT -------------------------------------------------------------- >5646_5646_52_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376139_length(amino acids)=400AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPL TGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFM -------------------------------------------------------------- >5646_5646_53_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376142_length(amino acids)=432AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPENISVPPPSGAPPAPPLAPLLPVSTVIAAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQ PHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLTGFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYA -------------------------------------------------------------- >5646_5646_54_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376160_length(amino acids)=399AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSISIAPPPPPMPQLTPQIPLT GFVARVQENIADSPTPPPPPPPDDIPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFME -------------------------------------------------------------- >5646_5646_55_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376166_length(amino acids)=370AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKHGNNQPARTGTL SRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSSIGIPI AVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDDIPMFD DSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTGLFPGN -------------------------------------------------------------- >5646_5646_56_APP-ABI1_APP_chr21_27542882_ENST00000439274_ABI1_chr10_27066170_ENST00000376170_length(amino acids)=375AA_BP=19 MLPGLALLLLAAWTARALETVDIHKEKVARREIGILTTNKNTSRTHKIIAPANMERPVRYIRKPIDYTVLDDVGHGVKWLKAKHGNNQPA RTGTLSRTNPPTQKPPSPPMSGRGTLGRNTPYKTLEPVKPPTVPNDYMTSPARLGSQHSPGRTASLNQRPRTHSGSSGGSGSRENSGSSS IGIPIAVPTPSPPTIGPAPGSAPGSQYGTMTRQISRHNSTTSSTSSGGYRRTPSVTAQFSAQPHVNGGPLYSQNSIADSPTPPPPPPPDD IPMFDDSPPPPPPPPVDYEDEEAAVVQYNDPYADGDPAWAPKNYIEKVVAIYDYTKDKDDELSFMEGAIIYVIKKNDDGWYEGVCNRVTG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:27542882/chr10:27066170) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| APP | ABI1 |
| FUNCTION: Multifunctional adapter protein that binds to various membrane receptors, nuclear factors and signaling proteins to regulate many processes, such as cell proliferation, immune response, endosomal trafficking and cell metabolism (PubMed:26583432, PubMed:15016378, PubMed:24879834). Regulates signaling pathway leading to cell proliferation through interaction with RAB5A and subunits of the NuRD/MeCP1 complex (PubMed:15016378). Plays a role in immune response by modulating phagocytosis, inflammatory and innate immune responses. In macrophages, enhances Fc-gamma receptor-mediated phagocytosis through interaction with RAB31 leading to activation of PI3K/Akt signaling. In response to LPS, modulates inflammatory responses by playing a key role on the regulation of TLR4 signaling and in the nuclear translocation of RELA/NF-kappa-B p65 and the secretion of pro- and anti-inflammatory cytokines. Also functions as a negative regulator of innate immune response via inhibition of AKT1 signaling pathway by forming a complex with APPL1 and PIK3R1 (By similarity). Plays a role in endosomal trafficking of TGFBR1 from the endosomes to the nucleus (PubMed:26583432). Plays a role in cell metabolism by regulating adiponecting ans insulin signaling pathways and adaptative thermogenesis (PubMed:24879834) (By similarity). In muscle, negatively regulates adiponectin-simulated glucose uptake and fatty acid oxidation by inhibiting adiponectin signaling pathway through APPL1 sequestration thereby antagonizing APPL1 action (By similarity). In muscles, negativeliy regulates insulin-induced plasma membrane recruitment of GLUT4 and glucose uptake through interaction with TBC1D1 (PubMed:24879834). Plays a role in cold and diet-induced adaptive thermogenesis by activating ventromedial hypothalamus (VMH) neurons throught AMPK inhibition which enhances sympathetic outflow to subcutaneous white adipose tissue (sWAT), sWAT beiging and cold tolerance (By similarity). Also plays a role in other signaling pathways namely Wnt/beta-catenin, HGF and glucocorticoid receptor signaling (PubMed:19433865) (By similarity). Positive regulator of beta-catenin/TCF-dependent transcription through direct interaction with RUVBL2/reptin resulting in the relief of RUVBL2-mediated repression of beta-catenin/TCF target genes by modulating the interactions within the beta-catenin-reptin-HDAC complex (PubMed:19433865). May affect adult neurogenesis in hippocampus and olfactory system via regulating the sensitivity of glucocorticoid receptor. Required for fibroblast migration through HGF cell signaling (By similarity). {ECO:0000250|UniProtKB:Q8K3G9, ECO:0000269|PubMed:15016378, ECO:0000269|PubMed:19433865, ECO:0000269|PubMed:24879834, ECO:0000269|PubMed:26583432}. | FUNCTION: May act in negative regulation of cell growth and transformation by interacting with nonreceptor tyrosine kinases ABL1 and/or ABL2. May play a role in regulation of EGF-induced Erk pathway activation. Involved in cytoskeletal reorganization and EGFR signaling. Together with EPS8 participates in transduction of signals from Ras to Rac. In vitro, a trimeric complex of ABI1, EPS8 and SOS1 exhibits Rac specific guanine nucleotide exchange factor (GEF) activity and ABI1 seems to act as an adapter in the complex. Regulates ABL1/c-Abl-mediated phosphorylation of ENAH. Recruits WASF1 to lamellipodia and there seems to regulate WASF1 protein level. In brain, seems to regulate the dendritic outgrowth and branching as well as to determine the shape and number of synaptic contacts of developing neurons. {ECO:0000269|PubMed:11003655, ECO:0000269|PubMed:18328268}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000346832 | 2 | 12 | 260_418 | 112.0 | 497.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000359188 | 1 | 11 | 260_418 | 95.0 | 481.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376138 | 1 | 10 | 260_418 | 95.0 | 453.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376139 | 1 | 10 | 260_418 | 95.0 | 477.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376140 | 1 | 11 | 260_418 | 95.0 | 482.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376142 | 1 | 12 | 260_418 | 95.0 | 509.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376166 | 1 | 9 | 260_418 | 95.0 | 447.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376170 | 1 | 10 | 260_418 | 95.0 | 452.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000490841 | 0 | 7 | 260_418 | 0 | 330.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000346832 | 2 | 12 | 446_505 | 112.0 | 497.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000359188 | 1 | 11 | 446_505 | 95.0 | 481.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376138 | 1 | 10 | 446_505 | 95.0 | 453.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376139 | 1 | 10 | 446_505 | 95.0 | 477.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376140 | 1 | 11 | 446_505 | 95.0 | 482.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376142 | 1 | 12 | 446_505 | 95.0 | 509.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376166 | 1 | 9 | 446_505 | 95.0 | 447.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376170 | 1 | 10 | 446_505 | 95.0 | 452.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000490841 | 0 | 7 | 446_505 | 0 | 330.0 | Domain | SH3 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000490841 | 0 | 7 | 45_107 | 0 | 330.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000490841 | 0 | 7 | 18_79 | 0 | 330.0 | Region | Required for binding to WASF1 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 230_260 | 19.0 | 771.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 274_280 | 19.0 | 771.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 230_260 | 19.0 | 696.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 274_280 | 19.0 | 696.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 230_260 | 19.0 | 640.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 274_280 | 19.0 | 640.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 230_260 | 19.0 | 752.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 274_280 | 19.0 | 752.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 230_260 | 19.0 | 753.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 274_280 | 19.0 | 753.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 230_260 | 19.0 | 715.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 274_280 | 19.0 | 715.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 230_260 | 0 | 747.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 274_280 | 0 | 747.0 | Compositional bias | Note=Poly-Thr |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 28_189 | 19.0 | 771.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 374_565 | 19.0 | 771.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 28_189 | 19.0 | 696.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 374_565 | 19.0 | 696.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 28_189 | 19.0 | 640.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 374_565 | 19.0 | 640.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 28_189 | 19.0 | 752.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 374_565 | 19.0 | 752.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 28_189 | 19.0 | 753.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 374_565 | 19.0 | 753.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 28_189 | 19.0 | 715.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 374_565 | 19.0 | 715.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 28_189 | 0 | 747.0 | Domain | E1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 374_565 | 0 | 747.0 | Domain | E2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 344_365 | 19.0 | 771.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 724_734 | 19.0 | 771.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 344_365 | 19.0 | 696.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 724_734 | 19.0 | 696.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 344_365 | 19.0 | 640.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 724_734 | 19.0 | 640.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 344_365 | 19.0 | 752.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 724_734 | 19.0 | 752.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 344_365 | 19.0 | 753.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 724_734 | 19.0 | 753.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 344_365 | 19.0 | 715.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 724_734 | 19.0 | 715.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 344_365 | 0 | 747.0 | Motif | OX-2 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 724_734 | 0 | 747.0 | Motif | Note=Basolateral sorting signal |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 131_189 | 19.0 | 771.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 181_188 | 19.0 | 771.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 28_123 | 19.0 | 771.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 391_423 | 19.0 | 771.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 491_522 | 19.0 | 771.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 523_540 | 19.0 | 771.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 96_110 | 19.0 | 771.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 131_189 | 19.0 | 696.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 181_188 | 19.0 | 696.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 28_123 | 19.0 | 696.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 391_423 | 19.0 | 696.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 491_522 | 19.0 | 696.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 523_540 | 19.0 | 696.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 96_110 | 19.0 | 696.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 131_189 | 19.0 | 640.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 181_188 | 19.0 | 640.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 28_123 | 19.0 | 640.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 391_423 | 19.0 | 640.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 491_522 | 19.0 | 640.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 523_540 | 19.0 | 640.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 96_110 | 19.0 | 640.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 131_189 | 19.0 | 752.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 181_188 | 19.0 | 752.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 28_123 | 19.0 | 752.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 391_423 | 19.0 | 752.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 491_522 | 19.0 | 752.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 523_540 | 19.0 | 752.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 96_110 | 19.0 | 752.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 131_189 | 19.0 | 753.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 181_188 | 19.0 | 753.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 28_123 | 19.0 | 753.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 391_423 | 19.0 | 753.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 491_522 | 19.0 | 753.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 523_540 | 19.0 | 753.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 96_110 | 19.0 | 753.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 131_189 | 19.0 | 715.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 181_188 | 19.0 | 715.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 28_123 | 19.0 | 715.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 391_423 | 19.0 | 715.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 491_522 | 19.0 | 715.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 523_540 | 19.0 | 715.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 96_110 | 19.0 | 715.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 131_189 | 0 | 747.0 | Region | CuBD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 181_188 | 0 | 747.0 | Region | Note=Zinc-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 28_123 | 0 | 747.0 | Region | GFLD subdomain |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 391_423 | 0 | 747.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 491_522 | 0 | 747.0 | Region | Note=Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 523_540 | 0 | 747.0 | Region | Collagen-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 96_110 | 0 | 747.0 | Region | Heparin-binding |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 18_701 | 19.0 | 771.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 723_770 | 19.0 | 771.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 18_701 | 19.0 | 696.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 723_770 | 19.0 | 696.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 18_701 | 19.0 | 640.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 723_770 | 19.0 | 640.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 18_701 | 19.0 | 752.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 723_770 | 19.0 | 752.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 18_701 | 19.0 | 753.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 723_770 | 19.0 | 753.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 18_701 | 19.0 | 715.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 723_770 | 19.0 | 715.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 18_701 | 0 | 747.0 | Topological domain | Extracellular |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 723_770 | 0 | 747.0 | Topological domain | Cytoplasmic |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 702_722 | 19.0 | 771.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 702_722 | 19.0 | 696.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 702_722 | 19.0 | 640.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 702_722 | 19.0 | 752.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 702_722 | 19.0 | 753.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 702_722 | 19.0 | 715.0 | Transmembrane | Helical |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 702_722 | 0 | 747.0 | Transmembrane | Helical |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000346832 | 2 | 12 | 45_107 | 112.0 | 497.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000359188 | 1 | 11 | 45_107 | 95.0 | 481.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376138 | 1 | 10 | 45_107 | 95.0 | 453.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376139 | 1 | 10 | 45_107 | 95.0 | 477.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376140 | 1 | 11 | 45_107 | 95.0 | 482.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376142 | 1 | 12 | 45_107 | 95.0 | 509.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376166 | 1 | 9 | 45_107 | 95.0 | 447.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376170 | 1 | 10 | 45_107 | 95.0 | 452.0 | Domain | t-SNARE coiled-coil homology | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000346832 | 2 | 12 | 18_79 | 112.0 | 497.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000359188 | 1 | 11 | 18_79 | 95.0 | 481.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376138 | 1 | 10 | 18_79 | 95.0 | 453.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376139 | 1 | 10 | 18_79 | 95.0 | 477.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376140 | 1 | 11 | 18_79 | 95.0 | 482.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376142 | 1 | 12 | 18_79 | 95.0 | 509.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376166 | 1 | 9 | 18_79 | 95.0 | 447.0 | Region | Required for binding to WASF1 | |
| Tgene | ABI1 | chr21:27542882 | chr10:27066170 | ENST00000376170 | 1 | 10 | 18_79 | 95.0 | 452.0 | Region | Required for binding to WASF1 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ABI1 | ENAH, NCKAP1, EPS8L1, EPS8L2, EPS8L3, EPS8, ABL1, SOS1, NCK1, RYK, CACNA1A, CBL, NCK2, MBP, CDC123, CTTN, PIK3R1, PRKAA1, SRRM1, TCERG1, PRPF40A, APBB1, GAS7, WASF2, ZNF511, ODF3L2, UBXN11, VASP, DTNBP1, LHX4, PCMT1, BAIAP2, CYFIP1, CYFIP2, NCKAP1L, SF3A3, SSB, WASF1, WASL, MKI67, NHS, PPP2R1A, HOMER3, ABI2, RBM14, BRK1, NHSL1, KIAA1522, RAPH1, THOC6, Gtf2e2, Sass6, Nhsl1, TCF4, ZNF746, CDH1, IKBIP, AMZ1, WASF3, TIMM50, NHSL2, CGB2, COG6, CCDC22, C10orf88, CDH23, NXN, TRIM14, PCM1, ESR2, HRAS, EZR, LAMP1, ATG16L1, ACTC1, UL135, RPS6KA2, WWP2, SMARCD1, FBXL19, TTC23, FHL2, RCOR3, KIAA1217, FAM124B, ENKD1, TRIP10, VARS2, DTNB, EIF3H, HOMER1, HHEX, SORBS2, PRKAA2, RBM15, NFASC, HNRNPK, LMO1, C22orf15, CCDC57, MAB21L2, CCHCR1, PLEKHA4, IVNS1ABP, CORO1C, BCLAF1, PDCD4, PRPSAP1, AKAP12, SLC25A3, TUBB4B, TUBA1B, TRA2B, PRPS1, CMPK1, PRPS2, TUBB, SLC25A5, PRPSAP2, MYO1C, HSPA6, HSPA8, HSPB1, CAD, HSPA7, DPP7, HSD17B12, SLC25A11, ITGA3, ABI1, ESR1, KTN1, KIAA0408, MRFAP1, SRP68, SYNE2, MRFAP1L1, DST, SORBS1, TLN1, ACTN2, HULC, RIN3, OGT, C11orf52, KRAS, LAMTOR1, LCK, LYN, OCLN, RAB35, RAB9A, RHOB, STX4, STX7, ZFPL1, FXR1, SHANK3, TSC1, ARHGAP12, YWHAG, FGFR1OP2, CCR1, ACAA1, SPG11, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| APP | |
| ABI1 | 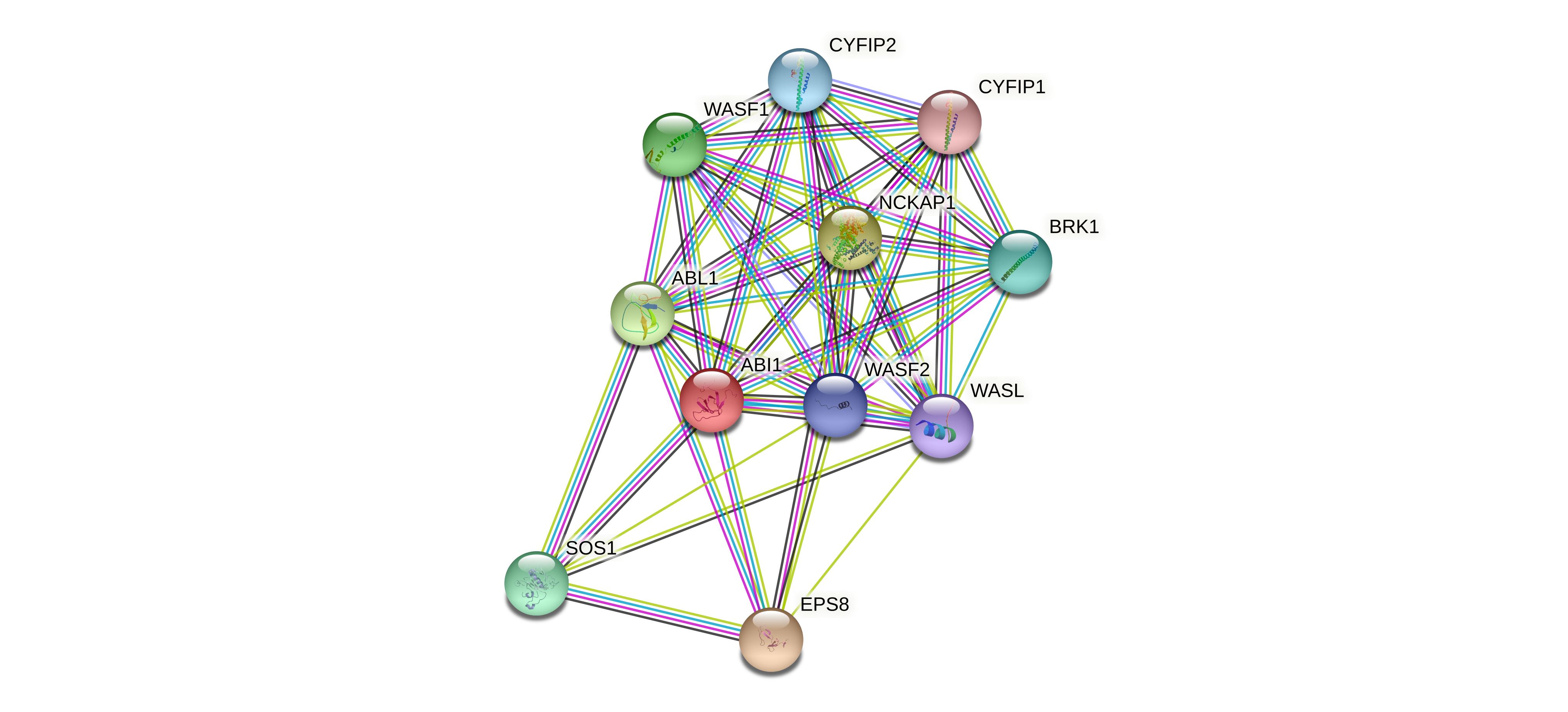 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 732_751 | 19.0 | 771.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 732_751 | 19.0 | 696.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 732_751 | 19.0 | 640.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 732_751 | 19.0 | 752.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 732_751 | 19.0 | 753.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 732_751 | 19.0 | 715.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 732_751 | 0 | 747.0 | G(o)-alpha |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000346798 | - | 1 | 18 | 695_722 | 19.0 | 771.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000348990 | - | 1 | 16 | 695_722 | 19.0 | 696.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000354192 | - | 1 | 15 | 695_722 | 19.0 | 640.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000357903 | - | 1 | 17 | 695_722 | 19.0 | 752.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000358918 | - | 1 | 17 | 695_722 | 19.0 | 753.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000359726 | - | 1 | 17 | 695_722 | 19.0 | 715.0 | PSEN1 |
| Hgene | APP | chr21:27542882 | chr10:27066170 | ENST00000440126 | - | 1 | 17 | 695_722 | 0 | 747.0 | PSEN1 |
Top |
Related Drugs to APP-ABI1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to APP-ABI1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | ABI1 | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Tgene | ABI1 | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Tgene | ABI1 | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Tgene | ABI1 | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Tgene | ABI1 | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Tgene | ABI1 | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |

