| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:NFE2L1-CARM1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: NFE2L1-CARM1 | FusionPDB ID: 58814 | FusionGDB2.0 ID: 58814 | Hgene | Tgene | Gene symbol | NFE2L1 | CARM1 | Gene ID | 4779 | 10498 |
| Gene name | nuclear factor, erythroid 2 like 1 | coactivator associated arginine methyltransferase 1 | |
| Synonyms | LCR-F1|NRF1|TCF11 | PRMT4 | |
| Cytomap | 17q21.32 | 19p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | endoplasmic reticulum membrane sensor NFE2L1NF-E2-related factor 1NFE2-related factor 1TCF-11locus control region-factor 1nuclear factor erythroid 2-related factor 1nuclear factor, erythroid derived 2, like 1protein NRF1, p120 formtranscription fa | histone-arginine methyltransferase CARM1protein arginine N-methyltransferase 4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q14494 | Q86X55 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000357480, ENST00000361665, ENST00000362042, ENST00000536222, ENST00000582155, ENST00000583378, ENST00000585291, ENST00000579481, | ENST00000327064, ENST00000344150, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 18 X 12 X 13=2808 | 4 X 7 X 10=280 |
| # samples | 20 | 12 | |
| ** MAII score | log2(20/2808*10)=-3.81147103052984 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/280*10)=-1.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: NFE2L1 [Title/Abstract] AND CARM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | NFE2L1(46134864)-CARM1(11022860), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | NFE2L1-CARM1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFE2L1-CARM1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFE2L1-CARM1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFE2L1-CARM1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CARM1 | GO:0016571 | histone methylation | 19405910 |
 Fusion gene breakpoints across NFE2L1 (5'-gene) Fusion gene breakpoints across NFE2L1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
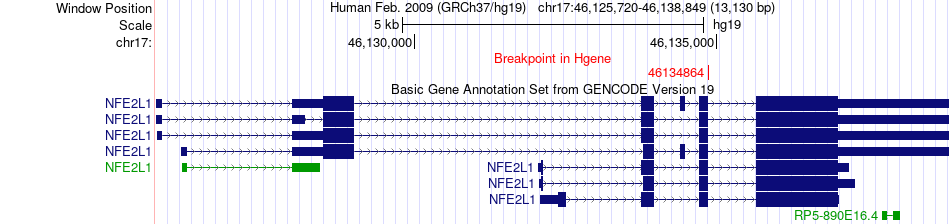 |
 Fusion gene breakpoints across CARM1 (3'-gene) Fusion gene breakpoints across CARM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
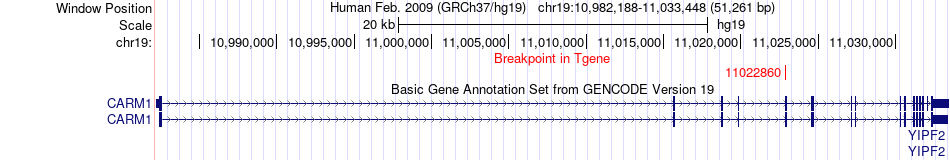 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-HC-7232-01A | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + |
| ChimerDB4 | PRAD | TCGA-HC-7232 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000362042 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 3872 | 1588 | 616 | 2856 | 746 |
| ENST00000362042 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 3752 | 1588 | 616 | 2787 | 723 |
| ENST00000585291 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 3481 | 1197 | 291 | 2465 | 724 |
| ENST00000585291 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 3361 | 1197 | 291 | 2396 | 701 |
| ENST00000357480 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 3771 | 1487 | 605 | 2755 | 716 |
| ENST00000357480 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 3651 | 1487 | 605 | 2686 | 693 |
| ENST00000361665 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 3827 | 1543 | 604 | 2811 | 735 |
| ENST00000361665 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 3707 | 1543 | 604 | 2742 | 712 |
| ENST00000582155 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 2743 | 459 | 51 | 1727 | 558 |
| ENST00000582155 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 2623 | 459 | 51 | 1658 | 535 |
| ENST00000583378 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 2693 | 409 | 34 | 1677 | 547 |
| ENST00000583378 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 2573 | 409 | 34 | 1608 | 524 |
| ENST00000536222 | NFE2L1 | chr17 | 46134864 | + | ENST00000327064 | CARM1 | chr19 | 11022860 | + | 3097 | 813 | 261 | 2081 | 606 |
| ENST00000536222 | NFE2L1 | chr17 | 46134864 | + | ENST00000344150 | CARM1 | chr19 | 11022860 | + | 2977 | 813 | 261 | 2012 | 583 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000362042 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001168036 | 0.9988319 |
| ENST00000362042 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001419519 | 0.99858046 |
| ENST00000585291 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001800093 | 0.9981998 |
| ENST00000585291 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.002192573 | 0.99780744 |
| ENST00000357480 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001166988 | 0.998833 |
| ENST00000357480 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001426954 | 0.99857306 |
| ENST00000361665 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001401241 | 0.99859875 |
| ENST00000361665 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.001584002 | 0.99841595 |
| ENST00000582155 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.002905432 | 0.9970945 |
| ENST00000582155 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.002426823 | 0.9975732 |
| ENST00000583378 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.004324501 | 0.9956755 |
| ENST00000583378 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.003678824 | 0.9963212 |
| ENST00000536222 | ENST00000327064 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.007771059 | 0.9922289 |
| ENST00000536222 | ENST00000344150 | NFE2L1 | chr17 | 46134864 | + | CARM1 | chr19 | 11022860 | + | 0.00791772 | 0.99208224 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >58814_58814_1_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000357480_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=716AA_BP=293 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLIDILWR QDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPALQNNLLSPL LTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEEVSLPEQVD IIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEYFRQPVVDT FDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQSPLFAKAGD TLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGMPTAYDLSS -------------------------------------------------------------- >58814_58814_2_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000357480_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=693AA_BP=293 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLIDILWR QDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPALQNNLLSPL LTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEEVSLPEQVD IIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEYFRQPVVDT FDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQSPLFAKAGD TLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGMPTAYDLSS -------------------------------------------------------------- >58814_58814_3_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000361665_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=735AA_BP=312 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLGAGREV FDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQVPSGEDQTALSLEECLRLLEATCPFGENAEFPADISSITE AVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTD RIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDL SALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLT HWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTY NLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNSQFTMGGPAISM -------------------------------------------------------------- >58814_58814_4_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000361665_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=712AA_BP=312 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLGAGREV FDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQVPSGEDQTALSLEECLRLLEATCPFGENAEFPADISSITE AVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTD RIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDL SALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLT HWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTY -------------------------------------------------------------- >58814_58814_5_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000362042_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=746AA_BP=323 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLIDILWR QDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQVPSGEDQTALSLEECLRLLEATCPFGENA EFPADISSITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHA EVLVKSNNLTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANF WYQPSFHGVDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTV WLSTAPTEPLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSP SENMWNTGSTYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNS -------------------------------------------------------------- >58814_58814_6_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000362042_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=723AA_BP=323 MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLIDILWR QDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQVPSGEDQTALSLEECLRLLEATCPFGENA EFPADISSITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHA EVLVKSNNLTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANF WYQPSFHGVDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTV WLSTAPTEPLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSP SENMWNTGSTYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLGSSGAQGSGGGSTSAHYAVNSQFTMGGPAISMASPMSIPTNTMH -------------------------------------------------------------- >58814_58814_7_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000536222_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=606AA_BP=183 MSGFPAPAALGLISQTMTRLDRSSHSGGGHAKGSRPLNSLLAGHRVAQPASAPGSRASVQDIDLIDILWRQDIDLGAGREVFDYSHRQKE QDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSI MEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESY LHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEA KEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISI VAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLANTG -------------------------------------------------------------- >58814_58814_8_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000536222_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=583AA_BP=183 MSGFPAPAALGLISQTMTRLDRSSHSGGGHAKGSRPLNSLLAGHRVAQPASAPGSRASVQDIDLIDILWRQDIDLGAGREVFDYSHRQKE QDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSI MEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESY LHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEA KEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISI VAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLGSSG -------------------------------------------------------------- >58814_58814_9_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000582155_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=558AA_BP=135 MGWESHLTAASADIDLIDILWRQDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISS ITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNN LTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHG VDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTE PLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTG STYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNSQFTMGGPA -------------------------------------------------------------- >58814_58814_10_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000582155_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=535AA_BP=135 MGWESHLTAASADIDLIDILWRQDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISS ITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNN LTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHG VDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTE PLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTG -------------------------------------------------------------- >58814_58814_11_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000583378_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=547AA_BP=124 MGWESHLTAASADIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEP PALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGK VEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAV DEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCL FQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAV AGMPTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNSQFTMGGPAISMASPMSIPT -------------------------------------------------------------- >58814_58814_12_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000583378_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=524AA_BP=124 MGWESHLTAASADIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEP PALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGK VEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAV DEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCL FQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAV -------------------------------------------------------------- >58814_58814_13_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000585291_CARM1_chr19_11022860_ENST00000327064_length(amino acids)=724AA_BP=301 MYVDCGETMLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARR LLSQVRALDRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDI DLIDILWRQDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPAL QNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEE VSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEY FRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQS PLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGM PTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNSQFTMGGPAISMASPMSIPTNTM -------------------------------------------------------------- >58814_58814_14_NFE2L1-CARM1_NFE2L1_chr17_46134864_ENST00000585291_CARM1_chr19_11022860_ENST00000344150_length(amino acids)=701AA_BP=301 MYVDCGETMLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARR LLSQVRALDRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDI DLIDILWRQDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQFPADISSITEAVPSESEPPAL QNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHAEVLVKSNNLTDRIVVIPGKVEE VSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANFWYQPSFHGVDLSALRGAAVDEY FRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTVWLSTAPTEPLTHWYQVRCLFQS PLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSPSENMWNTGSTYNLSSGMAVAGM -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:46134864/chr19:11022860) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFE2L1 | CARM1 |
| FUNCTION: [Endoplasmic reticulum membrane sensor NFE2L1]: Endoplasmic reticulum membrane sensor that translocates into the nucleus in response to various stresses to act as a transcription factor (PubMed:20932482, PubMed:24448410). Constitutes a precursor of the transcription factor NRF1 (By similarity). Able to detect various cellular stresses, such as cholesterol excess, oxidative stress or proteasome inhibition (PubMed:20932482). In response to stress, it is released from the endoplasmic reticulum membrane following cleavage by the protease DDI2 and translocates into the nucleus to form the transcription factor NRF1 (By similarity). Acts as a key sensor of cholesterol excess: in excess cholesterol conditions, the endoplasmic reticulum membrane form of the protein directly binds cholesterol via its CRAC motif, preventing cleavage and release of the transcription factor NRF1, thereby allowing expression of genes promoting cholesterol removal, such as CD36 (By similarity). Involved in proteasome homeostasis: in response to proteasome inhibition, it is released from the endoplasmic reticulum membrane, translocates to the nucleus and activates expression of genes encoding proteasome subunits (PubMed:20932482). {ECO:0000250|UniProtKB:Q61985, ECO:0000269|PubMed:20932482, ECO:0000269|PubMed:24448410}.; FUNCTION: [Transcription factor NRF1]: CNC-type bZIP family transcription factor that translocates to the nucleus and regulates expression of target genes in response to various stresses (PubMed:8932385, PubMed:9421508). Heterodimerizes with small-Maf proteins (MAFF, MAFG or MAFK) and binds DNA motifs including the antioxidant response elements (AREs), which regulate expression of genes involved in oxidative stress response (PubMed:8932385, PubMed:9421508). Activates or represses expression of target genes, depending on the context (PubMed:8932385, PubMed:9421508). Plays a key role in cholesterol homeostasis by acting as a sensor of cholesterol excess: in low cholesterol conditions, translocates into the nucleus and represses expression of genes involved in defense against cholesterol excess, such as CD36 (By similarity). In excess cholesterol conditions, the endoplasmic reticulum membrane form of the protein directly binds cholesterol via its CRAC motif, preventing cleavage and release of the transcription factor NRF1, thereby allowing expression of genes promoting cholesterol removal (By similarity). Critical for redox balance in response to oxidative stress: acts by binding the AREs motifs on promoters and mediating activation of oxidative stress response genes, such as GCLC, GCLM, GSS, MT1 and MT2 (By similarity). Plays an essential role during fetal liver hematopoiesis: probably has a protective function against oxidative stress and is involved in lipid homeostasis in the liver (By similarity). Involved in proteasome homeostasis: in response to proteasome inhibition, mediates the 'bounce-back' of proteasome subunits by translocating into the nucleus and activating expression of genes encoding proteasome subunits (PubMed:20932482). Also involved in regulating glucose flux (By similarity). Together with CEBPB; represses expression of DSPP during odontoblast differentiation (PubMed:15308669). In response to ascorbic acid induction, activates expression of SP7/Osterix in osteoblasts. {ECO:0000250|UniProtKB:Q61985, ECO:0000269|PubMed:15308669, ECO:0000269|PubMed:20932482, ECO:0000269|PubMed:8932385, ECO:0000269|PubMed:9421508}. | FUNCTION: Methylates (mono- and asymmetric dimethylation) the guanidino nitrogens of arginyl residues in several proteins involved in DNA packaging, transcription regulation, pre-mRNA splicing, and mRNA stability. Recruited to promoters upon gene activation together with histone acetyltransferases from EP300/P300 and p160 families, methylates histone H3 at 'Arg-17' (H3R17me), forming mainly asymmetric dimethylarginine (H3R17me2a), leading to activate transcription via chromatin remodeling. During nuclear hormone receptor activation and TCF7L2/TCF4 activation, acts synergically with EP300/P300 and either one of the p160 histone acetyltransferases NCOA1/SRC1, NCOA2/GRIP1 and NCOA3/ACTR or CTNNB1/beta-catenin to activate transcription. During myogenic transcriptional activation, acts together with NCOA3/ACTR as a coactivator for MEF2C. During monocyte inflammatory stimulation, acts together with EP300/P300 as a coactivator for NF-kappa-B. Acts as coactivator for PPARG, promotes adipocyte differentiation and the accumulation of brown fat tissue. Plays a role in the regulation of pre-mRNA alternative splicing by methylation of splicing factors. Also seems to be involved in p53/TP53 transcriptional activation. Methylates EP300/P300, both at 'Arg-2142', which may loosen its interaction with NCOA2/GRIP1, and at 'Arg-580' and 'Arg-604' in the KIX domain, which impairs its interaction with CREB and inhibits CREB-dependent transcriptional activation. Also methylates arginine residues in RNA-binding proteins PABPC1, ELAVL1 and ELAV4, which may affect their mRNA-stabilizing properties and the half-life of their target mRNAs. {ECO:0000269|PubMed:16497732, ECO:0000269|PubMed:19405910}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 125_288 | 294.0 | 743.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 125_288 | 324.0 | 773.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 125_288 | 294.0 | 743.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 191_199 | 294.0 | 743.0 | Region | Cholesterol recognition/amino acid consensus (CRAC) region |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 191_199 | 324.0 | 773.0 | Region | Cholesterol recognition/amino acid consensus (CRAC) region |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 191_199 | 294.0 | 743.0 | Region | Cholesterol recognition/amino acid consensus (CRAC) region |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 7_24 | 294.0 | 743.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 7_24 | 324.0 | 773.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 7_24 | 294.0 | 743.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | CARM1 | chr17:46134864 | chr19:11022860 | ENST00000327064 | 3 | 16 | 499_608 | 186.0 | 609.0 | Region | Transactivation domain | |
| Tgene | CARM1 | chr17:46134864 | chr19:11022860 | ENST00000344150 | 3 | 15 | 499_608 | 186.0 | 586.0 | Region | Transactivation domain |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 496_517 | 294.0 | 743.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 496_517 | 324.0 | 773.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 496_517 | 294.0 | 743.0 | Compositional bias | Note=Poly-Ser |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 654_717 | 294.0 | 743.0 | Domain | bZIP |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 654_717 | 324.0 | 773.0 | Domain | bZIP |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 654_717 | 294.0 | 743.0 | Domain | bZIP |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 476_480 | 294.0 | 743.0 | Motif | Destruction motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 476_480 | 324.0 | 773.0 | Motif | Destruction motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 476_480 | 294.0 | 743.0 | Motif | Destruction motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 379_383 | 294.0 | 743.0 | Region | CPD |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 656_675 | 294.0 | 743.0 | Region | Basic motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000357480 | + | 4 | 5 | 682_696 | 294.0 | 743.0 | Region | Leucine-zipper |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 379_383 | 324.0 | 773.0 | Region | CPD |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 656_675 | 324.0 | 773.0 | Region | Basic motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000362042 | + | 5 | 6 | 682_696 | 324.0 | 773.0 | Region | Leucine-zipper |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 379_383 | 294.0 | 743.0 | Region | CPD |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 656_675 | 294.0 | 743.0 | Region | Basic motif |
| Hgene | NFE2L1 | chr17:46134864 | chr19:11022860 | ENST00000585291 | + | 5 | 6 | 682_696 | 294.0 | 743.0 | Region | Leucine-zipper |
| Tgene | CARM1 | chr17:46134864 | chr19:11022860 | ENST00000327064 | 3 | 16 | 146_453 | 186.0 | 609.0 | Domain | SAM-dependent MTase PRMT-type | |
| Tgene | CARM1 | chr17:46134864 | chr19:11022860 | ENST00000344150 | 3 | 15 | 146_453 | 186.0 | 586.0 | Domain | SAM-dependent MTase PRMT-type |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1505_NFE2L1_46134864_CARM1_11022860_ranked_0.pdb | NFE2L1 | 46134864 | 46134864 | ENST00000344150 | CARM1 | chr19 | 11022860 | + | MLSLKKYLTEGLLQFTILLSLIGVRVDVDTYLTSQLPPLREIILGPSSAYTQTQFHNLRNTLDGYGIHPKSIDLDNYFTARRLLSQVRAL DRFQVPTTEVNAWLVHRDPEGSVSGSQPNSGLALESSSGLQDVTGPDNGVRESETEQGFGEDLEDLGAVAPPVSGDLTKEDIDLIDILWR QDIDLGAGREVFDYSHRQKEQDVEKELRDGGEQDTWAGEGAEALARNLLVDGETGESFPAQVPSGEDQTALSLEECLRLLEATCPFGENA EFPADISSITEAVPSESEPPALQNNLLSPLLTGTESPFDLEQQWQDLMSIMEMQIVLDVGCGSGILSFFAAQAGARKIYAVEASTMAQHA EVLVKSNNLTDRIVVIPGKVEEVSLPEQVDIIISEPMGYMLFNERMLESYLHAKKYLKPSGNMFPTIGDVHLAPFTDEQLYMEQFTKANF WYQPSFHGVDLSALRGAAVDEYFRQPVVDTFDIRILMAKSVKYTVNFLEAKEGDLHRIEIPFKFHMLHSGLVHGLAFWFDVAFIGSIMTV WLSTAPTEPLTHWYQVRCLFQSPLFAKAGDTLSGTCLLIANKRQSYDISIVAQVDQTGSKSSNLLDLKNPFFRYTGTTPSPPPGSHYTSP SENMWNTGSTYNLSSGMAVAGMPTAYDLSSVIASGSSVGHNNLIPLANTGIVNHTHSRMGSIMSTGIVQGSSGAQGSGGGSTSAHYAVNS | 746 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
NFE2L1_pLDDT.png |
CARM1_pLDDT.png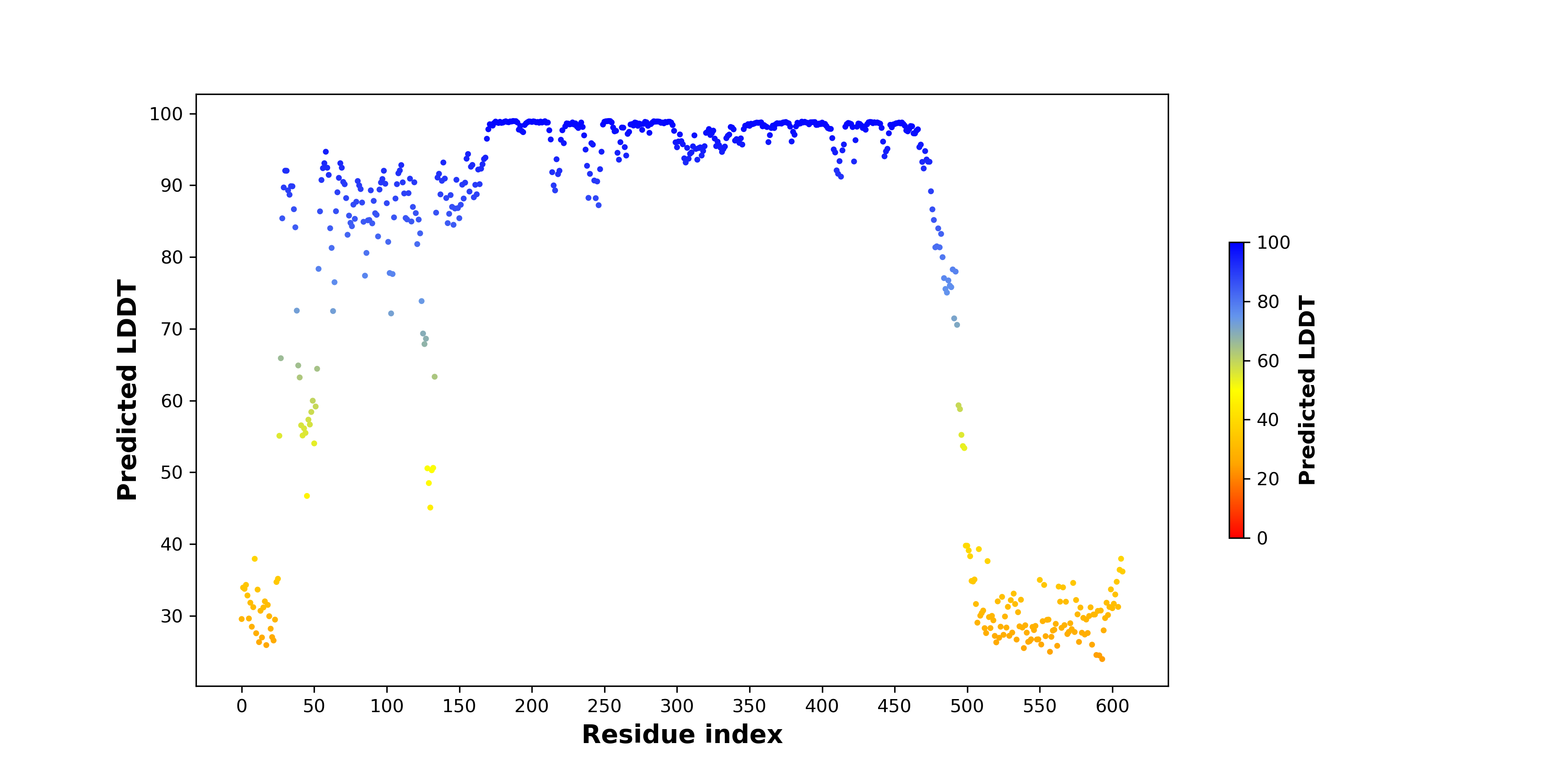 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| NFE2L1 | |
| CARM1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to NFE2L1-CARM1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to NFE2L1-CARM1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

