| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:NUMA1-RARA |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: NUMA1-RARA | FusionPDB ID: 60916 | FusionGDB2.0 ID: 60916 | Hgene | Tgene | Gene symbol | NUMA1 | RARA | Gene ID | 4926 | 5914 |
| Gene name | nuclear mitotic apparatus protein 1 | retinoic acid receptor alpha | |
| Synonyms | NMP-22|NUMA | NR1B1|RAR | |
| Cytomap | 11q13.4 | 17q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear mitotic apparatus protein 1SP-H antigencentrophilin stabilizes mitotic spindle in mitotic cellsnuclear matrix protein-22structural nuclear protein | retinoic acid receptor alphaRAR-alphanuclear receptor subfamily 1 group B member 1nucleophosmin-retinoic acid receptor alpha fusion protein NPM-RAR long formretinoic acid nuclear receptor alpha variant 1retinoic acid nuclear receptor alpha variant 2 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q14980 | P10276 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000351960, ENST00000358965, ENST00000393695, ENST00000543450, | ENST00000254066, ENST00000394081, ENST00000394086, ENST00000394089, ENST00000420042, ENST00000425707, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 36 X 22 X 17=13464 | 29 X 39 X 17=19227 |
| # samples | 43 | 55 | |
| ** MAII score | log2(43/13464*10)=-4.96862661174049 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(55/19227*10)=-5.12755824682814 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: NUMA1 [Title/Abstract] AND RARA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | NUMA1(71717270)-RARA(38504544), # samples:1 NUMA1(71717081)-RARA(38504544), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | NUMA1-RARA seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NUMA1-RARA seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NUMA1-RARA seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. NUMA1-RARA seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. NUMA1-RARA seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. NUMA1-RARA seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NUMA1 | GO:0000132 | establishment of mitotic spindle orientation | 21816348 |
| Hgene | NUMA1 | GO:0030953 | astral microtubule organization | 12445386 |
| Hgene | NUMA1 | GO:0060236 | regulation of mitotic spindle organization | 26195665 |
| Hgene | NUMA1 | GO:1902365 | positive regulation of protein localization to spindle pole body | 16076287 |
| Tgene | RARA | GO:0007165 | signal transduction | 2825025 |
| Tgene | RARA | GO:0030853 | negative regulation of granulocyte differentiation | 19917671 |
| Tgene | RARA | GO:0032689 | negative regulation of interferon-gamma production | 18416830 |
| Tgene | RARA | GO:0032720 | negative regulation of tumor necrosis factor production | 18416830 |
| Tgene | RARA | GO:0032736 | positive regulation of interleukin-13 production | 18416830 |
| Tgene | RARA | GO:0032753 | positive regulation of interleukin-4 production | 18416830 |
| Tgene | RARA | GO:0032754 | positive regulation of interleukin-5 production | 18416830 |
| Tgene | RARA | GO:0045630 | positive regulation of T-helper 2 cell differentiation | 18416830 |
| Tgene | RARA | GO:0045892 | negative regulation of transcription, DNA-templated | 20080953 |
| Tgene | RARA | GO:0045893 | positive regulation of transcription, DNA-templated | 18845237|19850744|20080953 |
| Tgene | RARA | GO:0045944 | positive regulation of transcription by RNA polymerase II | 19850744|21131358 |
| Tgene | RARA | GO:0071300 | cellular response to retinoic acid | 19917671 |
| Tgene | RARA | GO:0071391 | cellular response to estrogen stimulus | 20080953 |
 Fusion gene breakpoints across NUMA1 (5'-gene) Fusion gene breakpoints across NUMA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across RARA (3'-gene) Fusion gene breakpoints across RARA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | acute promyelocytic leukemia | AF012304 | NUMA1 | chr11 | 71717270 | RARA | chr17 | 38504544 | ||
| ChimerKB3 | . | . | NUMA1 | chr11 | 71717080 | - | RARA | chr17 | 38504567 | + |
| ChiTaRS5.0 | N/A | AF012304 | NUMA1 | chr11 | 71717081 | - | RARA | chr17 | 38504544 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000351960 | NUMA1 | chr11 | 71717080 | - | ENST00000254066 | RARA | chr17 | 38504567 | + | 4237 | 2456 | 154 | 3666 | 1170 |
| ENST00000351960 | NUMA1 | chr11 | 71717080 | - | ENST00000394089 | RARA | chr17 | 38504567 | + | 3772 | 2456 | 154 | 3666 | 1170 |
| ENST00000393695 | NUMA1 | chr11 | 71717080 | - | ENST00000254066 | RARA | chr17 | 38504567 | + | 7805 | 6024 | 314 | 7234 | 2306 |
| ENST00000393695 | NUMA1 | chr11 | 71717080 | - | ENST00000394089 | RARA | chr17 | 38504567 | + | 7340 | 6024 | 314 | 7234 | 2306 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >60916_60916_1_NUMA1-RARA_NUMA1_chr11_71717080_ENST00000351960_RARA_chr17_38504567_ENST00000254066_length(amino acids)=1170AA_BP=767 MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIHGTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSA QKVLEGSELELAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLNEDLENFLQKAPVPSTCSSTFPEELSPPSHQ AKREIRFLELQKVASSSSGNNFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLLTEKDAQIAMMQQRIDRLALL NEKQAASPLEPKELEELRDKNESLTMRLHETLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNELTEEHSKATQE WLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHLSQLQDNPPQEKGEVLGDVLQVEELSKKLADSDQASKVQQQKLKAVQAQGG ESQQEAQRLQAQLNELQAQLSQKEQAAEHYKLQMEKAKTHYDAKKQQNQELQEQLRSLEQLQKENKELRAEAERLGHELQQAGLKTKEAE QTCRHLTAQVRSLEAQVAHADQQLRDLGKFQVATDALKSREPQAKPQLDLSIDSLDLSCEEGTPLSITSKLPRTQPDGTSVPGEPASPIS QRLPPKVESLESLYFTPIPARSQAPLESSLDSLGDVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDSANSSFYSTRSAPASQASLRA TSSTQSLARLGSPDYGNSALLSLPGYRPTTRSSARRSQAGVSSGAPPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGV SACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFEVGMSKESVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVR KAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQD TMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDRVDMLQEPLLEALKVYVRKRRPSRPH MFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQPGGGGRDGGGLAPPPGSCSPSLSPSSNRSSPATHSP -------------------------------------------------------------- >60916_60916_2_NUMA1-RARA_NUMA1_chr11_71717080_ENST00000351960_RARA_chr17_38504567_ENST00000394089_length(amino acids)=1170AA_BP=767 MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIHGTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSA QKVLEGSELELAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLNEDLENFLQKAPVPSTCSSTFPEELSPPSHQ AKREIRFLELQKVASSSSGNNFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLLTEKDAQIAMMQQRIDRLALL NEKQAASPLEPKELEELRDKNESLTMRLHETLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNELTEEHSKATQE WLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHLSQLQDNPPQEKGEVLGDVLQVEELSKKLADSDQASKVQQQKLKAVQAQGG ESQQEAQRLQAQLNELQAQLSQKEQAAEHYKLQMEKAKTHYDAKKQQNQELQEQLRSLEQLQKENKELRAEAERLGHELQQAGLKTKEAE QTCRHLTAQVRSLEAQVAHADQQLRDLGKFQVATDALKSREPQAKPQLDLSIDSLDLSCEEGTPLSITSKLPRTQPDGTSVPGEPASPIS QRLPPKVESLESLYFTPIPARSQAPLESSLDSLGDVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDSANSSFYSTRSAPASQASLRA TSSTQSLARLGSPDYGNSALLSLPGYRPTTRSSARRSQAGVSSGAPPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGV SACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFEVGMSKESVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVR KAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQD TMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDRVDMLQEPLLEALKVYVRKRRPSRPH MFPKMLMKITDLRSISAKGAERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQPGGGGRDGGGLAPPPGSCSPSLSPSSNRSSPATHSP -------------------------------------------------------------- >60916_60916_3_NUMA1-RARA_NUMA1_chr11_71717080_ENST00000393695_RARA_chr17_38504567_ENST00000254066_length(amino acids)=2306AA_BP=1903 MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIHGTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSA QKVLEGSELELAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLNEDLENFLQKAPVPSTCSSTFPEELSPPSHQ AKREIRFLELQKVASSSSGNNFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLLTEKDAQIAMMQQRIDRLALL NEKQAASPLEPKELEELRDKNESLTMRLHETLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNELTEEHSKATQE WLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHLSQLQDNPPQEKGEVLGDVLQLETLKQEAATLAANNTQLQARVEMLETERG QQEAKLLAERGHFEEEKQQLSSLITDLQSSISNLSQAKEELEQASQAHGARLTAQVASLTSELTTLNATIQQQDQELAGLKQQAKEKQAQ LAQTLQQQEQASQGLRHQVEQLSSSLKQKEQQLKEVAEKQEATRQDHAQQLATAAEEREASLRERDAALKQLEALEKEKAAKLEILQQQL QVANEARDSAQTSVTQAQREKAELSRKVEELQACVETARQEQHEAQAQVAELELQLRSEQQKATEKERVAQEKDQLQEQLQALKESLKVT KGSLEEEKRRAADALEEQQRCISELKAETRSLVEQHKRERKELEEERAGRKGLEARLQQLGEAHQAETEVLRRELAEAMAAQHTAESECE QLVKEVAAWRERYEDSQQEEAQYGAMFQEQLMTLKEECEKARQELQEAKEKVAGIESHSELQISRQQNELAELHANLARALQQVQEKEVR AQKLADDLSTLQEKMAATSKEVARLETLVRKAGEQQETASRELVKEPARAGDRQPEWLEEQQGRQFCSTQAALQAMEREAEQMGNELERL RAALMESQGQQQEERGQQEREVARLTQERGRAQADLALEKAARAELEMRLQNALNEQRVEFATLQEALAHALTEKEGKDQELAKLRGLEA AQIKELEELRQTVKQLKEQLAKKEKEHASGSGAQSEAAGRTEPTGPKLEALRAEVSKLEQQCQKQQEQADSLERSLEAERASRAERDSAL ETLQGQLEEKAQELGHSQSALASAQRELAAFRTKVQDHSKAEDEWKAQVARGRQEAERKNSLISSLEEEVSILNRQVLEKEGESKELKRL VMAESEKSQKLEERLRLLQAETASNSARAAERSSALREEVQSLREEAEKQRVASENLRQELTSQAERAEELGQELKAWQEKFFQKEQALS TLQLEHTSTQALVSELLPAKHLCQQLQAEQAAAEKRHREELEQSKQAAGGLRAELLRAQRELGELIPLRQKVAEQERTAQQLRAEKASYA EQLSMLKKAHGLLAEENRGLGERANLGRQFLEVELDQAREKYVQELAAVRADAETRLAEVQREAQSTARELEVMTAKYEGAKVKVLEERQ RFQEERQKLTAQVEQLEVFQREQTKQVEELSKKLADSDQASKVQQQKLKAVQAQGGESQQEAQRLQAQLNELQAQLSQKEQAAEHYKLQM EKAKTHYDAKKQQNQELQEQLRSLEQLQKENKELRAEAERLGHELQQAGLKTKEAEQTCRHLTAQVRSLEAQVAHADQQLRDLGKFQVAT DALKSREPQAKPQLDLSIDSLDLSCEEGTPLSITSKLPRTQPDGTSVPGEPASPISQRLPPKVESLESLYFTPIPARSQAPLESSLDSLG DVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDSANSSFYSTRSAPASQASLRATSSTQSLARLGSPDYGNSALLSLPGYRPTTRSSA RRSQAGVSSGAPPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVT RNRCQYCRLQKCFEVGMSKESVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWD KFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQL LPLEMDDAETGLLSAICLICGDRQDLEQPDRVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSM -------------------------------------------------------------- >60916_60916_4_NUMA1-RARA_NUMA1_chr11_71717080_ENST00000393695_RARA_chr17_38504567_ENST00000394089_length(amino acids)=2306AA_BP=1903 MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIHGTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSA QKVLEGSELELAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLNEDLENFLQKAPVPSTCSSTFPEELSPPSHQ AKREIRFLELQKVASSSSGNNFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLLTEKDAQIAMMQQRIDRLALL NEKQAASPLEPKELEELRDKNESLTMRLHETLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNELTEEHSKATQE WLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHLSQLQDNPPQEKGEVLGDVLQLETLKQEAATLAANNTQLQARVEMLETERG QQEAKLLAERGHFEEEKQQLSSLITDLQSSISNLSQAKEELEQASQAHGARLTAQVASLTSELTTLNATIQQQDQELAGLKQQAKEKQAQ LAQTLQQQEQASQGLRHQVEQLSSSLKQKEQQLKEVAEKQEATRQDHAQQLATAAEEREASLRERDAALKQLEALEKEKAAKLEILQQQL QVANEARDSAQTSVTQAQREKAELSRKVEELQACVETARQEQHEAQAQVAELELQLRSEQQKATEKERVAQEKDQLQEQLQALKESLKVT KGSLEEEKRRAADALEEQQRCISELKAETRSLVEQHKRERKELEEERAGRKGLEARLQQLGEAHQAETEVLRRELAEAMAAQHTAESECE QLVKEVAAWRERYEDSQQEEAQYGAMFQEQLMTLKEECEKARQELQEAKEKVAGIESHSELQISRQQNELAELHANLARALQQVQEKEVR AQKLADDLSTLQEKMAATSKEVARLETLVRKAGEQQETASRELVKEPARAGDRQPEWLEEQQGRQFCSTQAALQAMEREAEQMGNELERL RAALMESQGQQQEERGQQEREVARLTQERGRAQADLALEKAARAELEMRLQNALNEQRVEFATLQEALAHALTEKEGKDQELAKLRGLEA AQIKELEELRQTVKQLKEQLAKKEKEHASGSGAQSEAAGRTEPTGPKLEALRAEVSKLEQQCQKQQEQADSLERSLEAERASRAERDSAL ETLQGQLEEKAQELGHSQSALASAQRELAAFRTKVQDHSKAEDEWKAQVARGRQEAERKNSLISSLEEEVSILNRQVLEKEGESKELKRL VMAESEKSQKLEERLRLLQAETASNSARAAERSSALREEVQSLREEAEKQRVASENLRQELTSQAERAEELGQELKAWQEKFFQKEQALS TLQLEHTSTQALVSELLPAKHLCQQLQAEQAAAEKRHREELEQSKQAAGGLRAELLRAQRELGELIPLRQKVAEQERTAQQLRAEKASYA EQLSMLKKAHGLLAEENRGLGERANLGRQFLEVELDQAREKYVQELAAVRADAETRLAEVQREAQSTARELEVMTAKYEGAKVKVLEERQ RFQEERQKLTAQVEQLEVFQREQTKQVEELSKKLADSDQASKVQQQKLKAVQAQGGESQQEAQRLQAQLNELQAQLSQKEQAAEHYKLQM EKAKTHYDAKKQQNQELQEQLRSLEQLQKENKELRAEAERLGHELQQAGLKTKEAEQTCRHLTAQVRSLEAQVAHADQQLRDLGKFQVAT DALKSREPQAKPQLDLSIDSLDLSCEEGTPLSITSKLPRTQPDGTSVPGEPASPISQRLPPKVESLESLYFTPIPARSQAPLESSLDSLG DVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDSANSSFYSTRSAPASQASLRATSSTQSLARLGSPDYGNSALLSLPGYRPTTRSSA RRSQAGVSSGAPPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVT RNRCQYCRLQKCFEVGMSKESVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWD KFSELSTKCIIKTVEFAKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLTDLVFAFANQL LPLEMDDAETGLLSAICLICGDRQDLEQPDRVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSM -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:71717270/chr17:38504544) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NUMA1 | RARA |
| FUNCTION: Microtubule (MT)-binding protein that plays a role in the formation and maintenance of the spindle poles and the alignement and the segregation of chromosomes during mitotic cell division (PubMed:7769006, PubMed:17172455, PubMed:19255246, PubMed:24996901, PubMed:26195665, PubMed:27462074). Functions to tether the minus ends of MTs at the spindle poles, which is critical for the establishment and maintenance of the spindle poles (PubMed:12445386, PubMed:11956313). Plays a role in the establishment of the mitotic spindle orientation during metaphase and elongation during anaphase in a dynein-dynactin-dependent manner (PubMed:23870127, PubMed:24109598, PubMed:24996901, PubMed:26765568). In metaphase, part of a ternary complex composed of GPSM2 and G(i) alpha proteins, that regulates the recruitment and anchorage of the dynein-dynactin complex in the mitotic cell cortex regions situated above the two spindle poles, and hence regulates the correct oritentation of the mitotic spindle (PubMed:23027904, PubMed:22327364, PubMed:23921553). During anaphase, mediates the recruitment and accumulation of the dynein-dynactin complex at the cell membrane of the polar cortical region through direct association with phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), and hence participates in the regulation of the spindle elongation and chromosome segregation (PubMed:22327364, PubMed:23921553, PubMed:24996901, PubMed:24371089). Binds also to other polyanionic phosphoinositides, such as phosphatidylinositol 3-phosphate (PIP), lysophosphatidic acid (LPA) and phosphatidylinositol triphosphate (PIP3), in vitro (PubMed:24996901, PubMed:24371089). Also required for proper orientation of the mitotic spindle during asymmetric cell divisions (PubMed:21816348). Plays a role in mitotic MT aster assembly (PubMed:11163243, PubMed:11229403, PubMed:12445386). Involved in anastral spindle assembly (PubMed:25657325). Positively regulates TNKS protein localization to spindle poles in mitosis (PubMed:16076287). Highly abundant component of the nuclear matrix where it may serve a non-mitotic structural role, occupies the majority of the nuclear volume (PubMed:10075938). Required for epidermal differentiation and hair follicle morphogenesis (By similarity). {ECO:0000250|UniProtKB:E9Q7G0, ECO:0000269|PubMed:11163243, ECO:0000269|PubMed:11229403, ECO:0000269|PubMed:11956313, ECO:0000269|PubMed:12445386, ECO:0000269|PubMed:16076287, ECO:0000269|PubMed:17172455, ECO:0000269|PubMed:19255246, ECO:0000269|PubMed:22327364, ECO:0000269|PubMed:23027904, ECO:0000269|PubMed:23870127, ECO:0000269|PubMed:23921553, ECO:0000269|PubMed:24109598, ECO:0000269|PubMed:24371089, ECO:0000269|PubMed:24996901, ECO:0000269|PubMed:25657325, ECO:0000269|PubMed:26195665, ECO:0000269|PubMed:26765568, ECO:0000269|PubMed:27462074, ECO:0000269|PubMed:7769006, ECO:0000305|PubMed:10075938, ECO:0000305|PubMed:21816348}. | FUNCTION: Receptor for retinoic acid (PubMed:19850744, PubMed:16417524, PubMed:20215566). Retinoic acid receptors bind as heterodimers to their target response elements in response to their ligands, all-trans or 9-cis retinoic acid, and regulate gene expression in various biological processes (PubMed:28167758). The RXR/RAR heterodimers bind to the retinoic acid response elements (RARE) composed of tandem 5'-AGGTCA-3' sites known as DR1-DR5 (PubMed:28167758, PubMed:19398580). In the absence of ligand, the RXR-RAR heterodimers associate with a multiprotein complex containing transcription corepressors that induce histone deacetylation, chromatin condensation and transcriptional suppression (PubMed:16417524). On ligand binding, the corepressors dissociate from the receptors and associate with the coactivators leading to transcriptional activation (PubMed:9267036, PubMed:19850744, PubMed:20215566). Formation of a complex with histone deacetylases might lead to inhibition of RARE DNA element binding and to transcriptional repression (PubMed:28167758). Transcriptional activation and RARE DNA element binding might be supported by the transcription factor KLF2 (PubMed:28167758). RARA plays an essential role in the regulation of retinoic acid-induced germ cell development during spermatogenesis (By similarity). Has a role in the survival of early spermatocytes at the beginning prophase of meiosis (By similarity). In Sertoli cells, may promote the survival and development of early meiotic prophase spermatocytes (By similarity). In concert with RARG, required for skeletal growth, matrix homeostasis and growth plate function (By similarity). Together with RXRA, positively regulates microRNA-10a expression, thereby inhibiting the GATA6/VCAM1 signaling response to pulsatile shear stress in vascular endothelial cells (PubMed:28167758). In association with HDAC3, HDAC5 and HDAC7 corepressors, plays a role in the repression of microRNA-10a and thereby promotes the inflammatory response (PubMed:28167758). {ECO:0000250|UniProtKB:P11416, ECO:0000269|PubMed:16417524, ECO:0000269|PubMed:19398580, ECO:0000269|PubMed:19850744, ECO:0000269|PubMed:20215566, ECO:0000269|PubMed:28167758, ECO:0000269|PubMed:9267036}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (839) >>>839.pdbFusion protein BP residue: 767 CIF file (839) >>>839.cif | NUMA1 | chr11 | 71717080 | - | RARA | chr17 | 38504567 | + | MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIH GTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSAQKVLEGSELE LAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLN EDLENFLQKAPVPSTCSSTFPEELSPPSHQAKREIRFLELQKVASSSSGN NFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLL TEKDAQIAMMQQRIDRLALLNEKQAASPLEPKELEELRDKNESLTMRLHE TLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNEL TEEHSKATQEWLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHL SQLQDNPPQEKGEVLGDVLQVEELSKKLADSDQASKVQQQKLKAVQAQGG ESQQEAQRLQAQLNELQAQLSQKEQAAEHYKLQMEKAKTHYDAKKQQNQE LQEQLRSLEQLQKENKELRAEAERLGHELQQAGLKTKEAEQTCRHLTAQV RSLEAQVAHADQQLRDLGKFQVATDALKSREPQAKPQLDLSIDSLDLSCE EGTPLSITSKLPRTQPDGTSVPGEPASPISQRLPPKVESLESLYFTPIPA RSQAPLESSLDSLGDVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDS ANSSFYSTRSAPASQASLRATSSTQSLARLGSPDYGNSALLSLPGYRPTT RSSARRSQAGVSSGAPPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQ DKSSGYHYGVSACEGCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQY CRLQKCFEVGMSKESVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVR KAHQETFPALCQLGKYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEF AKQLPGFTTLTIADQITLLKAACLDILILRICTRYTPEQDTMTFSDGLTL NRTQMHNAGFGPLTDLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDL EQPDRVDMLQEPLLEALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGA ERVITLKMEIPGSMPPLIQEMLENSEGLDTLSGQPGGGGRDGGGLAPPPG | 1170 |
| 3D view using mol* of 839 (AA BP:767) | ||||||||||
 | ||||||||||
| PDB file (1126) | NUMA1 | chr11 | 71717080 | - | RARA | chr17 | 38504567 | + | MSGITKMTLHATRGAALLSWVNSLHVADPVEAVLQLQDCSIFIKIIDRIH GTEEGQQILKQPVSERLDFVCSFLQKNRKHPSSPECLVSAQKVLEGSELE LAKMTMLLLYHSTMSSKSPRDWEQFEYKIQAELAVILKFVLDHEDGLNLN EDLENFLQKAPVPSTCSSTFPEELSPPSHQAKREIRFLELQKVASSSSGN NFLSGSPASPMGDILQTPQFQMRRLKKQLADERSNRDELELELAENRKLL TEKDAQIAMMQQRIDRLALLNEKQAASPLEPKELEELRDKNESLTMRLHE TLKQCQDLKTEKSQMDRKINQLSEENGDLSFKLREFASHLQQLQDALNEL TEEHSKATQEWLEKQAQLEKELSAALQDKKCLEEKNEILQGKLSQLEEHL SQLQDNPPQEKGEVLGDVLQLETLKQEAATLAANNTQLQARVEMLETERG QQEAKLLAERGHFEEEKQQLSSLITDLQSSISNLSQAKEELEQASQAHGA RLTAQVASLTSELTTLNATIQQQDQELAGLKQQAKEKQAQLAQTLQQQEQ ASQGLRHQVEQLSSSLKQKEQQLKEVAEKQEATRQDHAQQLATAAEEREA SLRERDAALKQLEALEKEKAAKLEILQQQLQVANEARDSAQTSVTQAQRE KAELSRKVEELQACVETARQEQHEAQAQVAELELQLRSEQQKATEKERVA QEKDQLQEQLQALKESLKVTKGSLEEEKRRAADALEEQQRCISELKAETR SLVEQHKRERKELEEERAGRKGLEARLQQLGEAHQAETEVLRRELAEAMA AQHTAESECEQLVKEVAAWRERYEDSQQEEAQYGAMFQEQLMTLKEECEK ARQELQEAKEKVAGIESHSELQISRQQNELAELHANLARALQQVQEKEVR AQKLADDLSTLQEKMAATSKEVARLETLVRKAGEQQETASRELVKEPARA GDRQPEWLEEQQGRQFCSTQAALQAMEREAEQMGNELERLRAALMESQGQ QQEERGQQEREVARLTQERGRAQADLALEKAARAELEMRLQNALNEQRVE FATLQEALAHALTEKEGKDQELAKLRGLEAAQIKELEELRQTVKQLKEQL AKKEKEHASGSGAQSEAAGRTEPTGPKLEALRAEVSKLEQQCQKQQEQAD SLERSLEAERASRAERDSALETLQGQLEEKAQELGHSQSALASAQRELAA FRTKVQDHSKAEDEWKAQVARGRQEAERKNSLISSLEEEVSILNRQVLEK EGESKELKRLVMAESEKSQKLEERLRLLQAETASNSARAAERSSALREEV QSLREEAEKQRVASENLRQELTSQAERAEELGQELKAWQEKFFQKEQALS TLQLEHTSTQALVSELLPAKHLCQQLQAEQAAAEKRHREELEQSKQAAGG LRAELLRAQRELGELIPLRQKVAEQERTAQQLRAEKASYAEQLSMLKKAH GLLAEENRGLGERANLGRQFLEVELDQAREKYVQELAAVRADAETRLAEV QREAQSTARELEVMTAKYEGAKVKVLEERQRFQEERQKLTAQVEQLEVFQ REQTKQVEELSKKLADSDQASKVQQQKLKAVQAQGGESQQEAQRLQAQLN ELQAQLSQKEQAAEHYKLQMEKAKTHYDAKKQQNQELQEQLRSLEQLQKE NKELRAEAERLGHELQQAGLKTKEAEQTCRHLTAQVRSLEAQVAHADQQL RDLGKFQVATDALKSREPQAKPQLDLSIDSLDLSCEEGTPLSITSKLPRT QPDGTSVPGEPASPISQRLPPKVESLESLYFTPIPARSQAPLESSLDSLG DVFLDSGRKTRSARRRTTQIINITMTKKLDVEEPDSANSSFYSTRSAPAS QASLRATSSTQSLARLGSPDYGNSALLSLPGYRPTTRSSARRSQAGVSSG APPAIETQSSSSEEIVPSPPSPPPLPRIYKPCFVCQDKSSGYHYGVSACE GCKGFFRRSIQKNMVYTCHRDKNCIINKVTRNRCQYCRLQKCFEVGMSKE SVRNDRNKKKKEVPKPECSESYTLTPEVGELIEKVRKAHQETFPALCQLG KYTTNNSSEQRVSLDIDLWDKFSELSTKCIIKTVEFAKQLPGFTTLTIAD QITLLKAACLDILILRICTRYTPEQDTMTFSDGLTLNRTQMHNAGFGPLT DLVFAFANQLLPLEMDDAETGLLSAICLICGDRQDLEQPDRVDMLQEPLL EALKVYVRKRRPSRPHMFPKMLMKITDLRSISAKGAERVITLKMEIPGSM PPLIQEMLENSEGLDTLSGQPGGGGRDGGGLAPPPGSCSPSLSPSSNRSS | 2306 |
Top |
pLDDT score distribution |
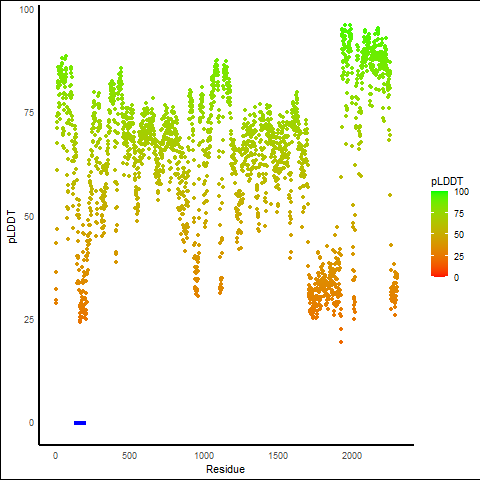
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
NUMA1_pLDDT.png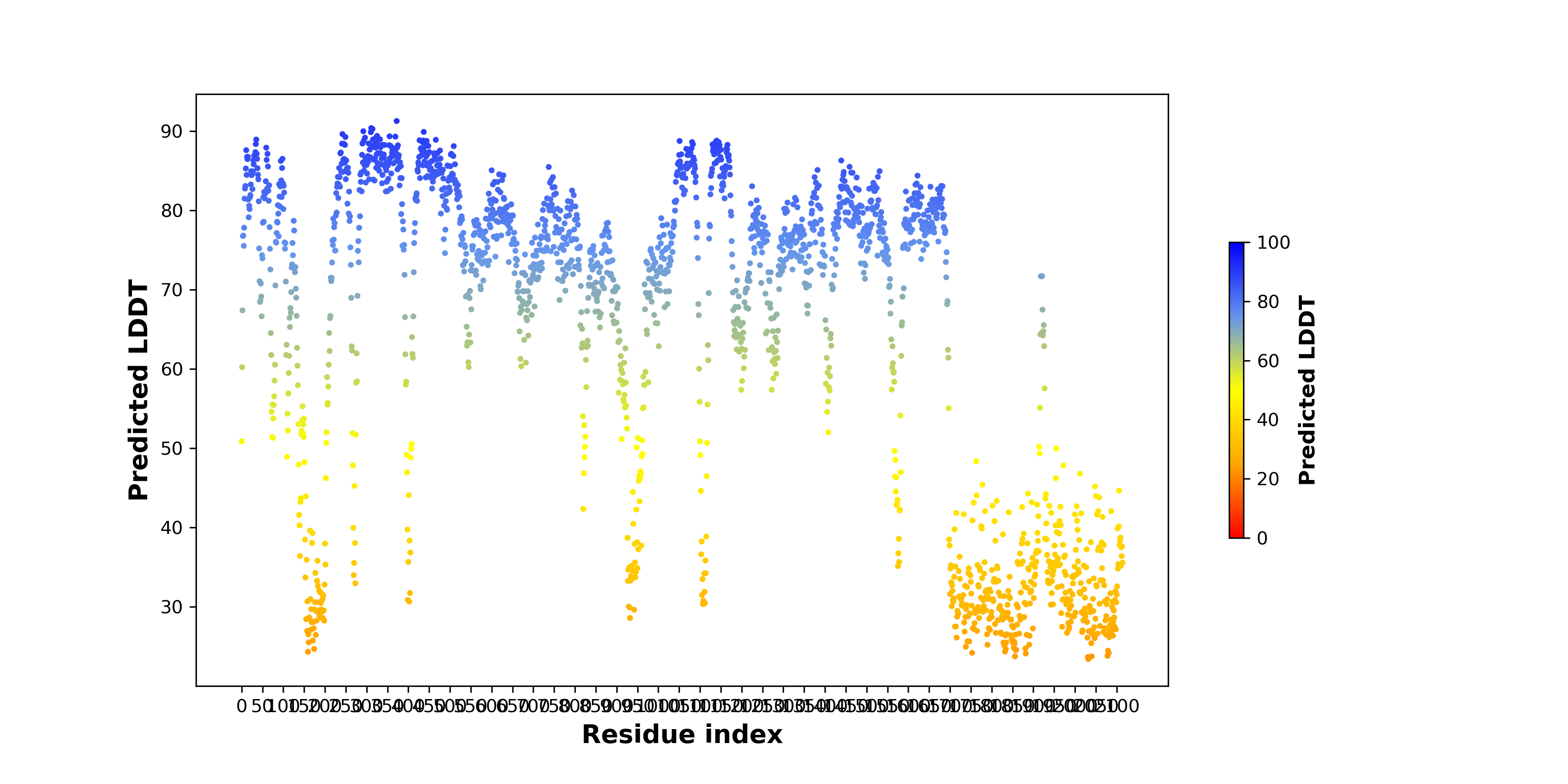 |
RARA_pLDDT.png |
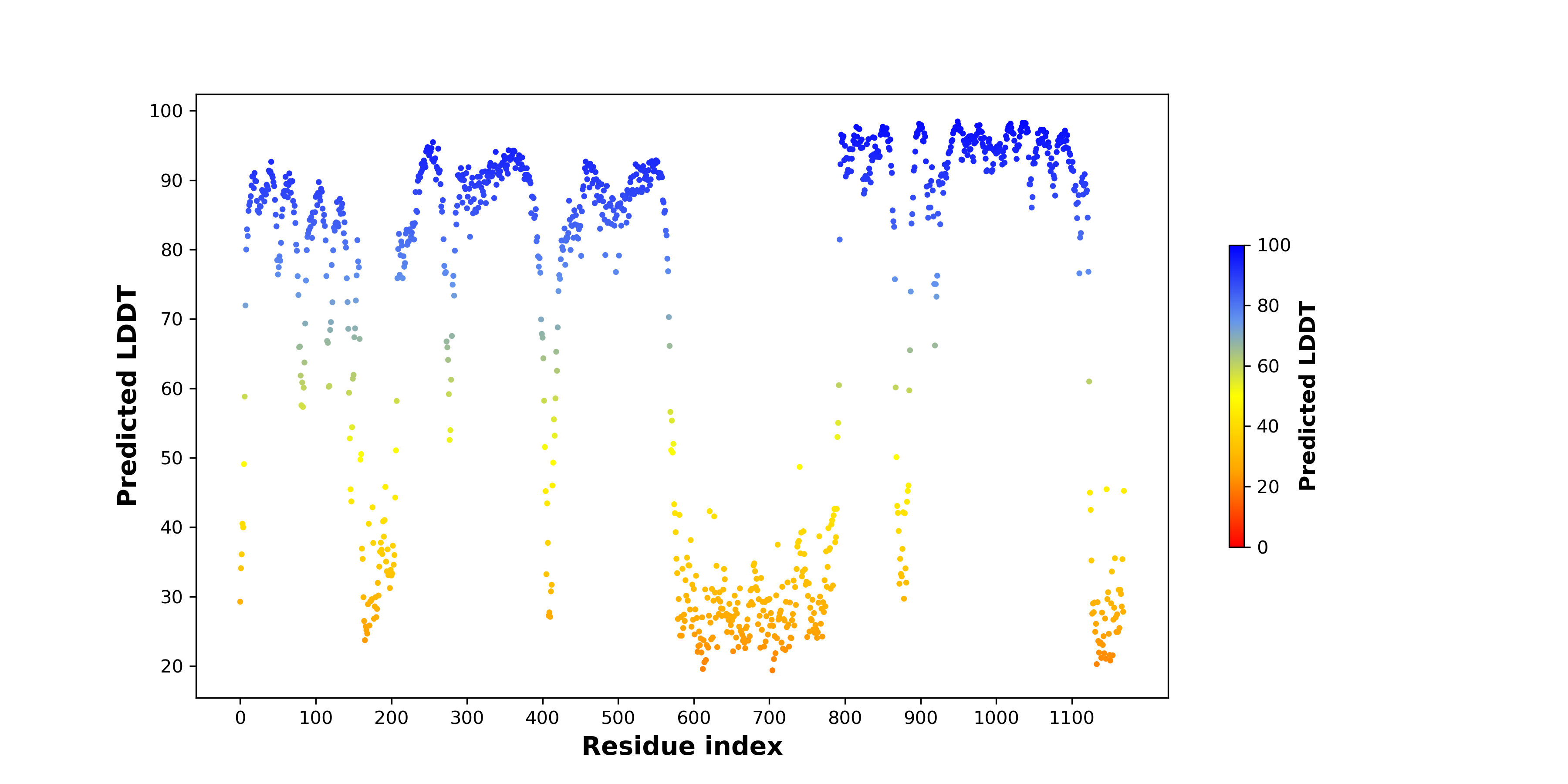
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
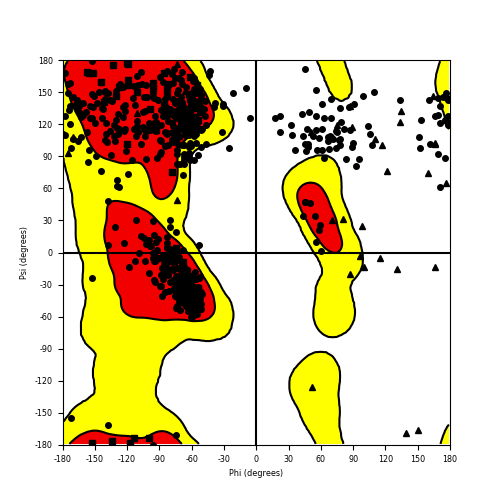
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| NUMA1_RARA_839.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 839 | 1.001 | 374 | 1.01 | 883.911 | 0.55 | 0.7 | 0.925 | 0.675 | 1.075 | 0.628 | 0.783 | Chain A: 20,46,49,50,54,69,72,73,74,76,77,78,79,80 ,83,85,87,107,110,111,112,113,114,115,116,117,118, 119,120,121,122,140,144,145,146,147,148,149,150,15 1,186,187,188,189,190,191,192,193,194,195,215,216, 218,220,221,223,224,225,227,228,229,231,232,233 |
| 1126 | 1.068 | 135 | 1.158 | 276.115 | 0.565 | 0.656 | 0.791 | 1.561 | 0.539 | 2.895 | 1.304 | Chain A: 131,134,135,137,138,141,142,144,145,149,1 50,151,153,154,157,158,159,185,187,188,190,191,193 ,194,195 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NUMA1 | EPB41, EPB41L1, YEATS4, GPSM2, TNKS, ACTR1A, SMC1A, SMC3, EPB41L2, NCOA6, PIM1, RPAP1, NPM1, BANF1, tat, HDAC5, Ranbp2, SMARCAD1, SIRT7, TERF1, TERF2IP, TERF2, CDK2, EMILIN1, RAE1, NUP98, DYNLL1, NCSTN, DDB1, ESR1, ECT2, MDC1, TP53BP1, SMURF1, VCP, HSP90AB1, HSP90AA1, CDK6, CBX6, KIAA0101, CCDC57, LMNA, YWHAQ, RPA3, RPA2, RPA1, ERG, LGR4, BRAP, DVL1, DVL2, DVL3, GMCL1, ENO1, MOV10, NXF1, BRCA1, CUL7, OBSL1, CCDC8, EZH2, SUZ12, EED, RNF2, BMI1, SUMO2, ABCE1, WDR83, GPSM1, EPB41L3, BRD4, EIF3H, CDC73, EIF3E, EIF3I, EIF3L, RPS13, SRRM1, RPL35A, RTF1, RUVBL1, ZC3H18, NTRK1, SCARNA22, PTTG1, IFI16, SSX2IP, CRK, MATR3, KIF20A, MPHOSPH8, CBX8, PAPD5, ASPM, Cbx1, Tnks, Srp72, Tnks2, DYNC1H1, KPNB1, FAM175B, BRCC3, U2AF2, SCOC, TCP10L, PDHA1, YAP1, TXNIP, MED12, CTNNB1, TCF7L2, EFTUD2, CHD3, CHD4, AGR2, MYC, CDK9, WDR76, HIST1H4A, HIST1H2BB, HIST1H2AB, HIST1H3A, KIAA1429, ACTC1, Bach1, CCND1, NR2C2, PPP1CC, ZFYVE21, XRCC6, DCAF15, BMH1, BMH2, ALYREF, SCARB2, FUS, EWSR1, TAF15, HMGB1, LRWD1, BIRC3, STAU1, WWP2, TRIM28, PLEKHA4, PINK1, WHSC1, FANCD2, GOLGA6L2, HIST1H1A, EIF5B, LYPLA2, NPR2, DDX10, LRRFIP1, RIBC1, HELZ2, MACF1, SDR42E2, VPS4B, MLYCD, LRPAP1, MMEL1, ZFYVE27, DMBX1, BIRC6, TLN1, PHIP, ORF14, LRRC31, CIT, ANLN, CHMP4B, KIF14, PRC1, PYHIN1, DEPDC1B, ARHGAP44, ARHGEF1, LRRC59, NDN, NUPR1, RBM39, nsp2, APEX1, ASF1A, HIST1H2BG, UBC, NAA40, ZBTB2, KRT4, KRT75, KRT72, KRT78, KXD1, TCP10, BTF3, EP300, FBXW7, nsp15, CCNB1, KPNA2, CCNF, TAX1BP1, SIRT6, YY1, PIGR, BRD2, BRD3, BRDT, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| NUMA1 | 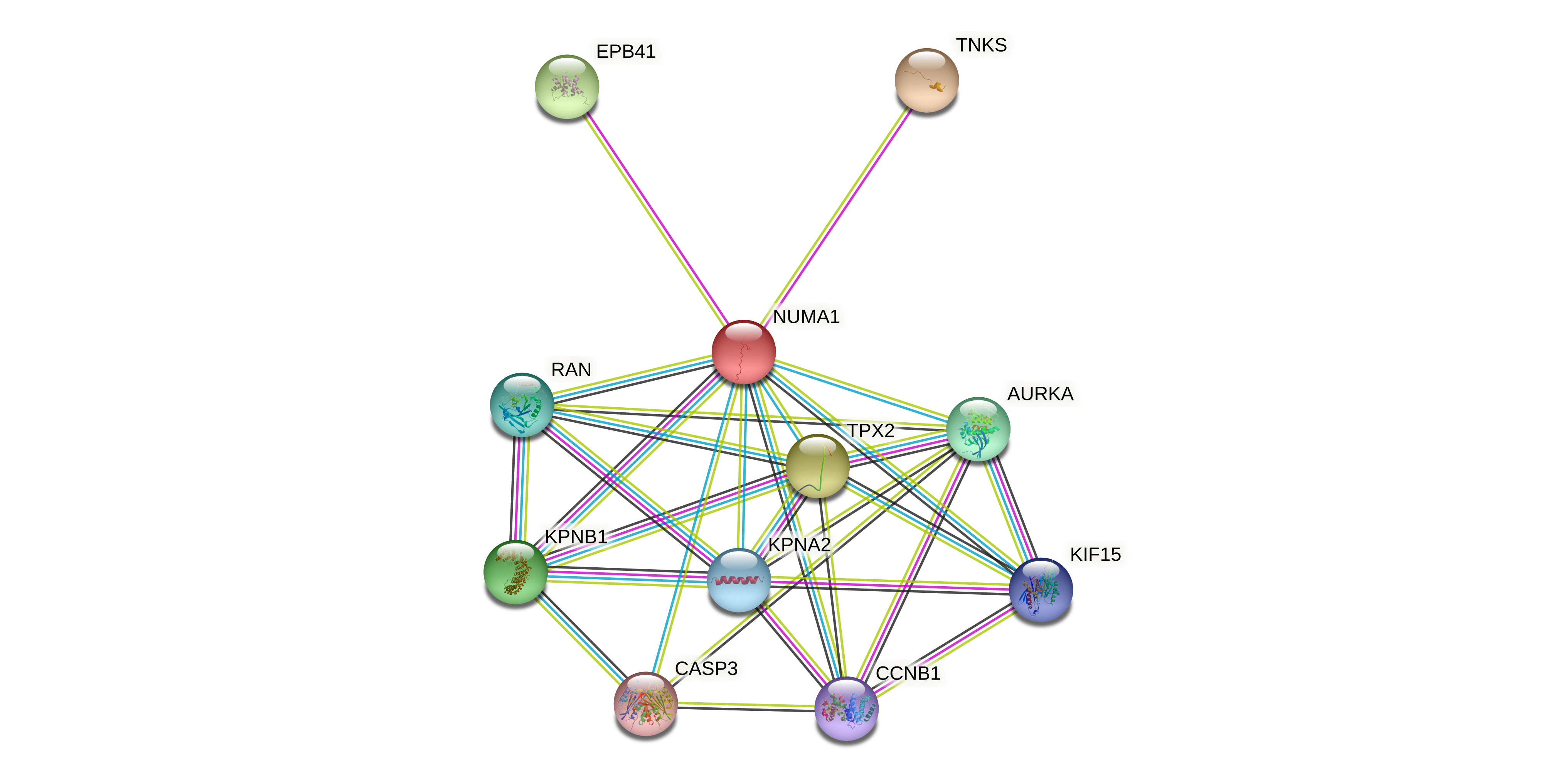 |
| RARA |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to NUMA1-RARA |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to NUMA1-RARA |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| NUMA1 | RARA | Invasive Breast Carcinoma | MyCancerGenome | |
| NUMA1 | RARA | Lung Adenocarcinoma | MyCancerGenome | |
| NUMA1 | RARA | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| NUMA1 | RARA | Esophageal Adenocarcinoma | MyCancerGenome | |
| NUMA1 | RARA | Acute Myeloid Leukemia | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | NUMA1 | C0023487 | Acute Promyelocytic Leukemia | 1 | CTD_human;ORPHANET |
| Hgene | NUMA1 | C0162820 | Dermatitis, Allergic Contact | 1 | CTD_human |
| Tgene | RARA | C0023487 | Acute Promyelocytic Leukemia | 24 | CTD_human;ORPHANET |
| Tgene | RARA | C0036341 | Schizophrenia | 3 | PSYGENET |
| Tgene | RARA | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | RARA | C0009363 | Congenital ocular coloboma (disorder) | 1 | GENOMICS_ENGLAND |
| Tgene | RARA | C0010701 | Phyllodes Tumor | 1 | CTD_human |
| Tgene | RARA | C0085183 | Neoplasms, Second Primary | 1 | CTD_human |
| Tgene | RARA | C0086696 | Neoplasms, Therapy-Associated | 1 | CTD_human |
| Tgene | RARA | C0149940 | Sciatic Neuropathy | 1 | CTD_human |
| Tgene | RARA | C0154748 | Lesion of Sciatic Nerve | 1 | CTD_human |
| Tgene | RARA | C0206650 | Fibroadenoma | 1 | CTD_human |
| Tgene | RARA | C0242013 | Sciatic Neuritis | 1 | CTD_human |
| Tgene | RARA | C0525045 | Mood Disorders | 1 | PSYGENET |
| Tgene | RARA | C0600066 | Malignant Cystosarcoma Phyllodes | 1 | CTD_human |
| Tgene | RARA | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | RARA | C0751924 | Neuralgia-Neuritis, Sciatic Nerve | 1 | CTD_human |
| Tgene | RARA | C0751925 | Sciatic Nerve Palsy | 1 | CTD_human |
| Tgene | RARA | C0877578 | Treatment related secondary malignancy | 1 | CTD_human |
| Tgene | RARA | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | RARA | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | RARA | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | RARA | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |