| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:RUNX1-USP42 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: RUNX1-USP42 | FusionPDB ID: 78697 | FusionGDB2.0 ID: 78697 | Hgene | Tgene | Gene symbol | RUNX1 | USP42 | Gene ID | 861 | 84132 |
| Gene name | RUNX family transcription factor 1 | ubiquitin specific peptidase 42 | |
| Synonyms | AML1|AML1-EVI-1|AMLCR1|CBF2alpha|CBFA2|EVI-1|PEBP2aB|PEBP2alpha | - | |
| Cytomap | 21q22.12 | 7p22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | runt-related transcription factor 1AML1-EVI-1 fusion proteinPEA2-alpha BPEBP2-alpha BSL3-3 enhancer factor 1 alpha B subunitSL3/AKV core-binding factor alpha B subunitacute myeloid leukemia 1 proteincore-binding factor, runt domain, alpha subunit 2 | ubiquitin carboxyl-terminal hydrolase 42deubiquitinating enzyme 42ubiquitin specific protease 42ubiquitin thioesterase 42ubiquitin thiolesterase 42ubiquitin-specific-processing protease 42 | |
| Modification date | 20200322 | 20200320 | |
| UniProtAcc | Q06455 | Q9H9J4 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000437180, ENST00000300305, ENST00000325074, ENST00000344691, ENST00000399240, ENST00000358356, ENST00000486278, ENST00000494829, | ENST00000306177, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 46 X 68 X 13=40664 | 6 X 8 X 7=336 |
| # samples | 89 | 8 | |
| ** MAII score | log2(89/40664*10)=-5.51380298959468 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/336*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: RUNX1 [Title/Abstract] AND USP42 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | RUNX1(36167758)-USP42(6150914), # samples:1 RUNX1(36167489)-USP42(6150914), # samples:1 USP42(6151080)-RUNX1(36167584), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RUNX1-USP42 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. RUNX1-USP42 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. RUNX1-USP42 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. RUNX1-USP42 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RUNX1 | GO:0030097 | hemopoiesis | 21873977 |
| Hgene | RUNX1 | GO:0045893 | positive regulation of transcription, DNA-templated | 10207087|14970218 |
| Hgene | RUNX1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 9199349|10207087|14970218|21873977 |
| Tgene | USP42 | GO:0016579 | protein deubiquitination | 14715245 |
 Fusion gene breakpoints across RUNX1 (5'-gene) Fusion gene breakpoints across RUNX1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across USP42 (3'-gene) Fusion gene breakpoints across USP42 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | acute myeloid leukemia | FR727326 | RUNX1 | chr21 | 36167758 | USP42 | chr7 | 6150914 | ||
| ChimerKB3 | . | . | RUNX1 | chr21 | 36171597 | - | USP42 | chr7 | 6154953 | + |
| ChiTaRS5.0 | N/A | FR727326 | RUNX1 | chr21 | 36167489 | - | USP42 | chr7 | 6150914 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000344691 | RUNX1 | chr21 | 36171597 | - | ENST00000306177 | USP42 | chr7 | 6154953 | + | 7220 | 2464 | 1578 | 6173 | 1531 |
| ENST00000325074 | RUNX1 | chr21 | 36171597 | - | ENST00000306177 | USP42 | chr7 | 6154953 | + | 5687 | 931 | 0 | 4640 | 1546 |
| ENST00000300305 | RUNX1 | chr21 | 36171597 | - | ENST00000306177 | USP42 | chr7 | 6154953 | + | 6168 | 1412 | 445 | 5121 | 1558 |
| ENST00000399240 | RUNX1 | chr21 | 36171597 | - | ENST00000306177 | USP42 | chr7 | 6154953 | + | 5521 | 765 | 71 | 4474 | 1467 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >78697_78697_1_RUNX1-USP42_RUNX1_chr21_36171597_ENST00000300305_USP42_chr7_6154953_ENST00000306177_length(amino acids)=1558AA_BP=316 MASDSIFESFPSYPQCFMRECILGMNPSRDVHDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPGELV RTDSPNFLCSVLPTHWRCNKTLPIAFKVVALGDVPDGTLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTITVF TNPPQVATYHRAIKITVDGPREPRRHRQKLDDQTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQPQSQMQDT RQIQPSPPWSYDQSYQYLGSIASPSVHPATPISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGN TCFANAALQCLTYTPPLANYMLSHEHSKTCHAEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDA MQKACLNGSNKLDRHTQATTLVCQIFGGYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMV PASKRFTIHRSSNVLTLSLKRFANFTGGKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMND SIVSTSDIRSVLSQQAYVLFYIRSHDVKNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGTGPLKDTPS SSMSSPNGNSSVNRASPVNASASVQNWSVNRSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTITNSAVQSTSN ASTMSVSSKVTKPIPRSESCSQPVMNGKSKLNSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLN GANSADSDSDPKENGLAPDGASCQGQPALHSENPFAKANGLPGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDA EPQPGSPAAESLEEPDAAAGLSSTKKAPPPRDPGTPATKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDAKGM IAEGPRDSALAEAPEGLSPAPPARSEEPCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSPGERVEDAA APKAPGPSPAKEKIGSLRKVDRGHYRSRRERSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSPGERRSLGRCS HHHSRHRSGVELDWVRHHYTEGERGWGREKFYPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARE RERHRPSSPRAGAPHALAPHPDRFSHDRTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKS KKKKKSKDKHRDRDSRHQQDSDLSAACSDADLHRHKKKKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPLEGV -------------------------------------------------------------- >78697_78697_2_RUNX1-USP42_RUNX1_chr21_36171597_ENST00000325074_USP42_chr7_6154953_ENST00000306177_length(amino acids)=1546AA_BP=304 MPAAPRGPAQGEAAARTRSRDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPGELVRTDSPNFLCSVL PTHWRCNKTLPIAFKVVALGDVPDGTLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRA IKITVDGPREPRRHRQKLDDQTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQPQSQMQDTRQIQPSPPWSYD QSYQYLGSIASPSVHPATPISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGNTCFANAALQCLT YTPPLANYMLSHEHSKTCHAEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDAMQKACLNGSNKL DRHTQATTLVCQIFGGYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMVPASKRFTIHRSS NVLTLSLKRFANFTGGKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMNDSIVSTSDIRSVL SQQAYVLFYIRSHDVKNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGTGPLKDTPSSSMSSPNGNSSV NRASPVNASASVQNWSVNRSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTITNSAVQSTSNASTMSVSSKVTK PIPRSESCSQPVMNGKSKLNSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLNGANSADSDSDPK ENGLAPDGASCQGQPALHSENPFAKANGLPGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDAEPQPGSPAAESL EEPDAAAGLSSTKKAPPPRDPGTPATKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDAKGMIAEGPRDSALAE APEGLSPAPPARSEEPCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSPGERVEDAAAPKAPGPSPAKE KIGSLRKVDRGHYRSRRERSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSPGERRSLGRCSHHHSRHRSGVEL DWVRHHYTEGERGWGREKFYPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARERERHRPSSPRAG APHALAPHPDRFSHDRTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKSKKKKKSKDKHRD RDSRHQQDSDLSAACSDADLHRHKKKKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPLEGVGPFREKTKHLRM -------------------------------------------------------------- >78697_78697_3_RUNX1-USP42_RUNX1_chr21_36171597_ENST00000344691_USP42_chr7_6154953_ENST00000306177_length(amino acids)=1531AA_BP=289 MRIPVDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPGELVRTDSPNFLCSVLPTHWRCNKTLPIAFK VVALGDVPDGTLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRAIKITVDGPREPRRHR QKLDDQTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQPQSQMQDTRQIQPSPPWSYDQSYQYLGSIASPSVH PATPISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGNTCFANAALQCLTYTPPLANYMLSHEHS KTCHAEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDAMQKACLNGSNKLDRHTQATTLVCQIFG GYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMVPASKRFTIHRSSNVLTLSLKRFANFTG GKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMNDSIVSTSDIRSVLSQQAYVLFYIRSHDV KNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGTGPLKDTPSSSMSSPNGNSSVNRASPVNASASVQNW SVNRSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTITNSAVQSTSNASTMSVSSKVTKPIPRSESCSQPVMNG KSKLNSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLNGANSADSDSDPKENGLAPDGASCQGQP ALHSENPFAKANGLPGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDAEPQPGSPAAESLEEPDAAAGLSSTKKA PPPRDPGTPATKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDAKGMIAEGPRDSALAEAPEGLSPAPPARSEE PCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSPGERVEDAAAPKAPGPSPAKEKIGSLRKVDRGHYRS RRERSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSPGERRSLGRCSHHHSRHRSGVELDWVRHHYTEGERGWG REKFYPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARERERHRPSSPRAGAPHALAPHPDRFSHD RTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKSKKKKKSKDKHRDRDSRHQQDSDLSAAC SDADLHRHKKKKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPLEGVGPFREKTKHLRMESRDDRCRLFEYGQG -------------------------------------------------------------- >78697_78697_4_RUNX1-USP42_RUNX1_chr21_36171597_ENST00000399240_USP42_chr7_6154953_ENST00000306177_length(amino acids)=1467AA_BP=225 MRIPVDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPGELVRTDSPNFLCSVLPTHWRCNKTLPIAFK VVALGDVPDGTLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRAIKITVDGPREPRNTR QIQPSPPWSYDQSYQYLGSIASPSVHPATPISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGNT CFANAALQCLTYTPPLANYMLSHEHSKTCHAEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDAM QKACLNGSNKLDRHTQATTLVCQIFGGYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMVP ASKRFTIHRSSNVLTLSLKRFANFTGGKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMNDS IVSTSDIRSVLSQQAYVLFYIRSHDVKNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGTGPLKDTPSS SMSSPNGNSSVNRASPVNASASVQNWSVNRSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTITNSAVQSTSNA STMSVSSKVTKPIPRSESCSQPVMNGKSKLNSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLNG ANSADSDSDPKENGLAPDGASCQGQPALHSENPFAKANGLPGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDAE PQPGSPAAESLEEPDAAAGLSSTKKAPPPRDPGTPATKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDAKGMI AEGPRDSALAEAPEGLSPAPPARSEEPCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSPGERVEDAAA PKAPGPSPAKEKIGSLRKVDRGHYRSRRERSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSPGERRSLGRCSH HHSRHRSGVELDWVRHHYTEGERGWGREKFYPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARER ERHRPSSPRAGAPHALAPHPDRFSHDRTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKSK KKKKSKDKHRDRDSRHQQDSDLSAACSDADLHRHKKKKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPLEGVG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:36167758/chr7:6150914) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| RUNX1 | USP42 |
| FUNCTION: Transcriptional corepressor which facilitates transcriptional repression via its association with DNA-binding transcription factors and recruitment of other corepressors and histone-modifying enzymes (PubMed:12559562, PubMed:15203199, PubMed:10688654). Can repress the expression of MMP7 in a ZBTB33-dependent manner (PubMed:23251453). Can repress transactivation mediated by TCF12 (PubMed:16803958). Acts as a negative regulator of adipogenesis (By similarity). The AML1-MTG8/ETO fusion protein frequently found in leukemic cells is involved in leukemogenesis and contributes to hematopoietic stem/progenitor cell self-renewal (PubMed:23812588). {ECO:0000250|UniProtKB:Q61909, ECO:0000269|PubMed:10688654, ECO:0000269|PubMed:10973986, ECO:0000269|PubMed:16803958, ECO:0000269|PubMed:23251453, ECO:0000269|PubMed:23812588, ECO:0000303|PubMed:12559562, ECO:0000303|PubMed:15203199}. | FUNCTION: Deubiquitinating enzyme which may play an important role during spermatogenesis. {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (936)CIF file (936) >>>936.cif | RUNX1 | chr21 | 36171597 | - | USP42 | chr7 | 6154953 | + | MRIPVDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRS MVEVLADHPGELVRTDSPNFLCSVLPTHWRCNKTLPIAFKVVALGDVPDG TLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTI TVFTNPPQVATYHRAIKITVDGPREPRNTRQIQPSPPWSYDQSYQYLGSI ASPSVHPATPISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKI CLKWQQTHRVGAGLQNLGNTCFANAALQCLTYTPPLANYMLSHEHSKTCH AEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAH EFLQYTVDAMQKACLNGSNKLDRHTQATTLVCQIFGGYLRSRVKCLNCKG VSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMVP ASKRFTIHRSSNVLTLSLKRFANFTGGKIAKDVKYPEYLDIRPYMSQPNG EPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQMNDSIVSTSDIRSV LSQQAYVLFYIRSHDVKNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAP GFIGPQLPSHMIKNPPHLNGTGPLKDTPSSSMSSPNGNSSVNRASPVNAS ASVQNWSVNRSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENP TKPVPSSTITNSAVQSTSNASTMSVSSKVTKPIPRSESCSQPVMNGKSKL NSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPH ELQEPMTLNGANSADSDSDPKENGLAPDGASCQGQPALHSENPFAKANGL PGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDAE PQPGSPAAESLEEPDAAAGLSSTKKAPPPRDPGTPATKEGAWEAMAVAPE EPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDAKGMIAEGPRDSALA EAPEGLSPAPPARSEEPCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERP PAPVLDMAPAGHPEGDAEPSPGERVEDAAAPKAPGPSPAKEKIGSLRKVD RGHYRSRRERSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHH PGHGDRLSPGERRSLGRCSHHHSRHRSGVELDWVRHHYTEGERGWGREKF YPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRG RRGCEPARERERHRPSSPRAGAPHALAPHPDRFSHDRTALVAGDNCNLSD RFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKSK KKKKSKDKHRDRDSRHQQDSDLSAACSDADLHRHKKKKKKKKRHSRKSED FVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPLEGVGPFREKTKHLR | 1467 |
| 3D view using mol* of 936 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (952)CIF file (952) >>>952.cif | RUNX1 | chr21 | 36171597 | - | USP42 | chr7 | 6154953 | + | MRIPVDASTSRRFTPPSTALSPGKMSEALPLGAPDAGAALAGKLRSGDRS MVEVLADHPGELVRTDSPNFLCSVLPTHWRCNKTLPIAFKVVALGDVPDG TLVTVMAGNDENYSAELRNATAAMKNQVARFNDLRFVGRSGRGKSFTLTI TVFTNPPQVATYHRAIKITVDGPREPRRHRQKLDDQTKPGSLSFSERLSE LEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQPQSQMQDTRQIQPSP PWSYDQSYQYLGSIASPSVHPATPISPGRASGMTTLSAELSSRLSTLGDG IAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGNTCFANAALQCLTYTPPL ANYMLSHEHSKTCHAEGFCMMCTMQAHITQALSNPGDVIKPMFVINEMRR IARHFRFGNQEDAHEFLQYTVDAMQKACLNGSNKLDRHTQATTLVCQIFG GYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSVNKALEQFVKPEQLDG ENSYKCSKCKKMVPASKRFTIHRSSNVLTLSLKRFANFTGGKIAKDVKYP EYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAGHYFCYIKASNGLWYQ MNDSIVSTSDIRSVLSQQAYVLFYIRSHDVKNGGELTHPTHSPGQSSPRP VISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGTGPLKDTPSSSMSSPN GNSSVNRASPVNASASVQNWSVNRSSVIPEHPKKQKITISIHNKLPVRQC QSQPNLHSNSLENPTKPVPSSTITNSAVQSTSNASTMSVSSKVTKPIPRS ESCSQPVMNGKSKLNSSVLVPYGAESSEDSDEESKGLGKENGIGTIVSSH SPGQDAEDEEATPHELQEPMTLNGANSADSDSDPKENGLAPDGASCQGQP ALHSENPFAKANGLPGKLMPAPLLSLPEDKILETFRLSNKLKGSTDEMSA PGAERGPPEDRDAEPQPGSPAAESLEEPDAAAGLSSTKKAPPPRDPGTPA TKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCDPGSLTGDASPLSQDA KGMIAEGPRDSALAEAPEGLSPAPPARSEEPCEQPLLVHPSGDHARDAQD PSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSPGERVEDAAAPKAPGP SPAKEKIGSLRKVDRGHYRSRRERSSSGEPARESRSKTEGHRHRRRRTCP RERDRQDRHAPEHHPGHGDRLSPGERRSLGRCSHHHSRHRSGVELDWVRH HYTEGERGWGREKFYPDRPRWDRCRYYHDRYALYAARDWKPFHGGREHER AGLHERPHKDHNRGRRGCEPARERERHRPSSPRAGAPHALAPHPDRFSHD RTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENSDSHVEKKARRSEQKD PLEEPKAKKHKKSKKKKKSKDKHRDRDSRHQQDSDLSAACSDADLHRHKK KKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQFRRAQGGFPLSGGPPL | 1531 |
| 3D view using mol* of 952 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (957)CIF file (957) >>>957.cif | RUNX1 | chr21 | 36171597 | - | USP42 | chr7 | 6154953 | + | MPAAPRGPAQGEAAARTRSRDASTSRRFTPPSTALSPGKMSEALPLGAPD AGAALAGKLRSGDRSMVEVLADHPGELVRTDSPNFLCSVLPTHWRCNKTL PIAFKVVALGDVPDGTLVTVMAGNDENYSAELRNATAAMKNQVARFNDLR FVGRSGRGKSFTLTITVFTNPPQVATYHRAIKITVDGPREPRRHRQKLDD QTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPRASLNHSTAFNPQ PQSQMQDTRQIQPSPPWSYDQSYQYLGSIASPSVHPATPISPGRASGMTT LSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHRVGAGLQNLGNTC FANAALQCLTYTPPLANYMLSHEHSKTCHAEGFCMMCTMQAHITQALSNP GDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDAMQKACLNGSNKL DRHTQATTLVCQIFGGYLRSRVKCLNCKGVSDTFDPYLDITLEIKAAQSV NKALEQFVKPEQLDGENSYKCSKCKKMVPASKRFTIHRSSNVLTLSLKRF ANFTGGKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYAVLVHTGFNCHAG HYFCYIKASNGLWYQMNDSIVSTSDIRSVLSQQAYVLFYIRSHDVKNGGE LTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPSHMIKNPPHLNGT GPLKDTPSSSMSSPNGNSSVNRASPVNASASVQNWSVNRSSVIPEHPKKQ KITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTITNSAVQSTSNAS TMSVSSKVTKPIPRSESCSQPVMNGKSKLNSSVLVPYGAESSEDSDEESK GLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLNGANSADSDSDPK ENGLAPDGASCQGQPALHSENPFAKANGLPGKLMPAPLLSLPEDKILETF RLSNKLKGSTDEMSAPGAERGPPEDRDAEPQPGSPAAESLEEPDAAAGLS STKKAPPPRDPGTPATKEGAWEAMAVAPEEPPPSAGEDIVGDTAPPDLCD PGSLTGDASPLSQDAKGMIAEGPRDSALAEAPEGLSPAPPARSEEPCEQP LLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAPAGHPEGDAEPSP GERVEDAAAPKAPGPSPAKEKIGSLRKVDRGHYRSRRERSSSGEPARESR SKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSPGERRSLGRCSHH HSRHRSGVELDWVRHHYTEGERGWGREKFYPDRPRWDRCRYYHDRYALYA ARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARERERHRPSSPRAG APHALAPHPDRFSHDRTALVAGDNCNLSDRFHEHENGKSRKRRHDSVENS DSHVEKKARRSEQKDPLEEPKAKKHKKSKKKKKSKDKHRDRDSRHQQDSD LSAACSDADLHRHKKKKKKKKRHSRKSEDFVKDSELHLPRVTSLETVAQF | 1546 |
| 3D view using mol* of 957 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (961)CIF file (961) >>>961.cif | RUNX1 | chr21 | 36171597 | - | USP42 | chr7 | 6154953 | + | MASDSIFESFPSYPQCFMRECILGMNPSRDVHDASTSRRFTPPSTALSPG KMSEALPLGAPDAGAALAGKLRSGDRSMVEVLADHPGELVRTDSPNFLCS VLPTHWRCNKTLPIAFKVVALGDVPDGTLVTVMAGNDENYSAELRNATAA MKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRAIKITVDGP REPRRHRQKLDDQTKPGSLSFSERLSELEQLRRTAMRVSPHHPAPTPNPR ASLNHSTAFNPQPQSQMQDTRQIQPSPPWSYDQSYQYLGSIASPSVHPAT PISPGRASGMTTLSAELSSRLSTLGDGIAPPQKVLFPSEKICLKWQQTHR VGAGLQNLGNTCFANAALQCLTYTPPLANYMLSHEHSKTCHAEGFCMMCT MQAHITQALSNPGDVIKPMFVINEMRRIARHFRFGNQEDAHEFLQYTVDA MQKACLNGSNKLDRHTQATTLVCQIFGGYLRSRVKCLNCKGVSDTFDPYL DITLEIKAAQSVNKALEQFVKPEQLDGENSYKCSKCKKMVPASKRFTIHR SSNVLTLSLKRFANFTGGKIAKDVKYPEYLDIRPYMSQPNGEPIVYVLYA VLVHTGFNCHAGHYFCYIKASNGLWYQMNDSIVSTSDIRSVLSQQAYVLF YIRSHDVKNGGELTHPTHSPGQSSPRPVISQRVVTNKQAAPGFIGPQLPS HMIKNPPHLNGTGPLKDTPSSSMSSPNGNSSVNRASPVNASASVQNWSVN RSSVIPEHPKKQKITISIHNKLPVRQCQSQPNLHSNSLENPTKPVPSSTI TNSAVQSTSNASTMSVSSKVTKPIPRSESCSQPVMNGKSKLNSSVLVPYG AESSEDSDEESKGLGKENGIGTIVSSHSPGQDAEDEEATPHELQEPMTLN GANSADSDSDPKENGLAPDGASCQGQPALHSENPFAKANGLPGKLMPAPL LSLPEDKILETFRLSNKLKGSTDEMSAPGAERGPPEDRDAEPQPGSPAAE SLEEPDAAAGLSSTKKAPPPRDPGTPATKEGAWEAMAVAPEEPPPSAGED IVGDTAPPDLCDPGSLTGDASPLSQDAKGMIAEGPRDSALAEAPEGLSPA PPARSEEPCEQPLLVHPSGDHARDAQDPSQSLGAPEAAERPPAPVLDMAP AGHPEGDAEPSPGERVEDAAAPKAPGPSPAKEKIGSLRKVDRGHYRSRRE RSSSGEPARESRSKTEGHRHRRRRTCPRERDRQDRHAPEHHPGHGDRLSP GERRSLGRCSHHHSRHRSGVELDWVRHHYTEGERGWGREKFYPDRPRWDR CRYYHDRYALYAARDWKPFHGGREHERAGLHERPHKDHNRGRRGCEPARE RERHRPSSPRAGAPHALAPHPDRFSHDRTALVAGDNCNLSDRFHEHENGK SRKRRHDSVENSDSHVEKKARRSEQKDPLEEPKAKKHKKSKKKKKSKDKH RDRDSRHQQDSDLSAACSDADLHRHKKKKKKKKRHSRKSEDFVKDSELHL PRVTSLETVAQFRRAQGGFPLSGGPPLEGVGPFREKTKHLRMESRDDRCR | 1558 |
| 3D view using mol* of 961 (AA BP:) | ||||||||||
 | ||||||||||
Top |
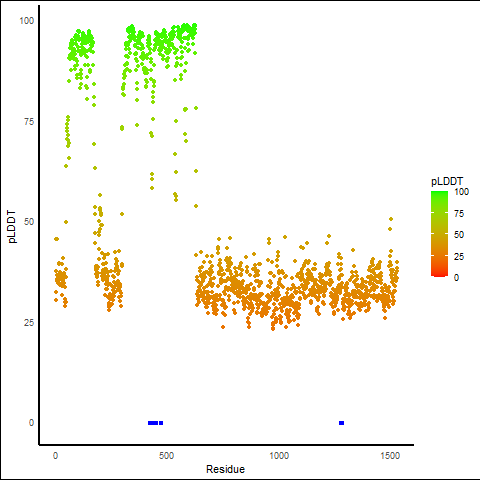
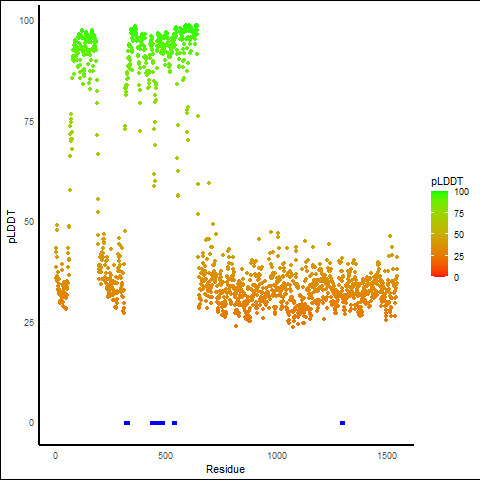
pLDDT score distribution |
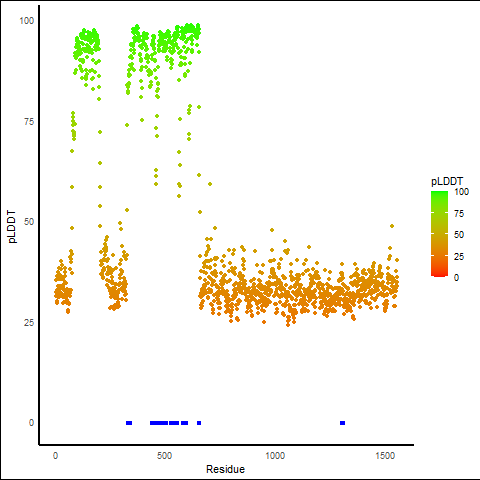
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
RUNX1_pLDDT.png |
USP42_pLDDT.png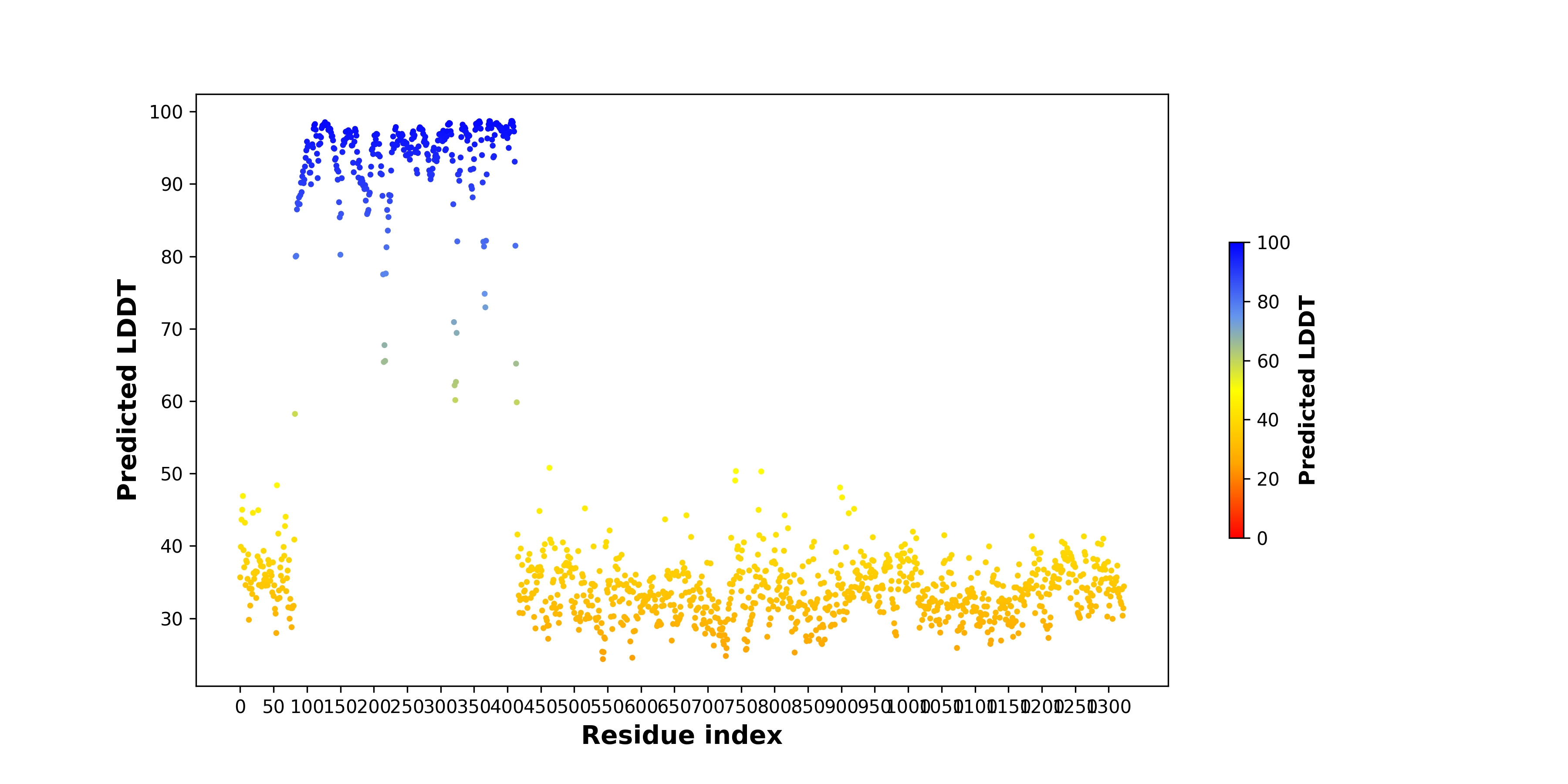 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
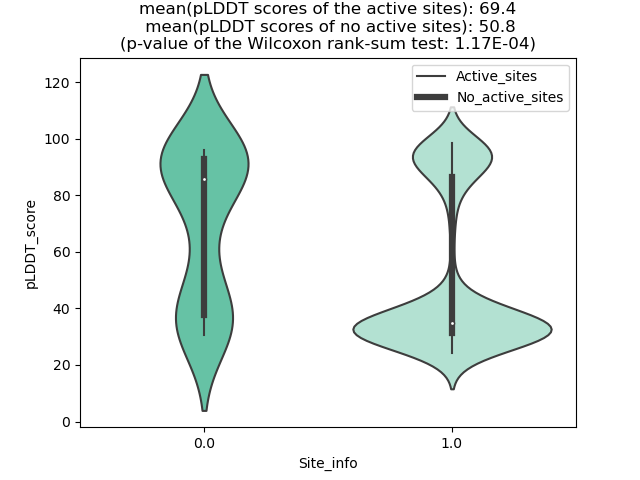
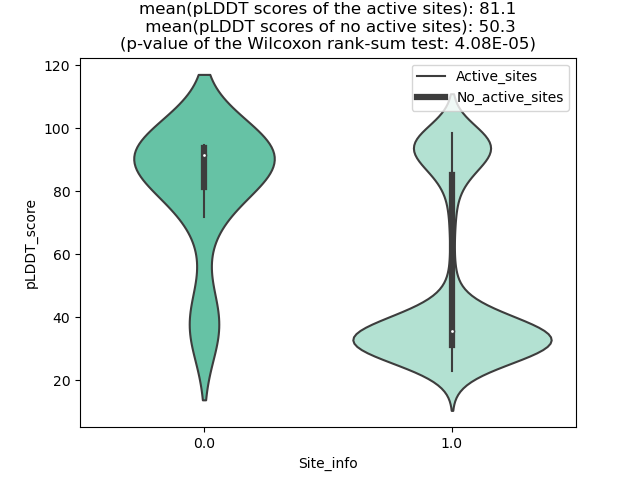
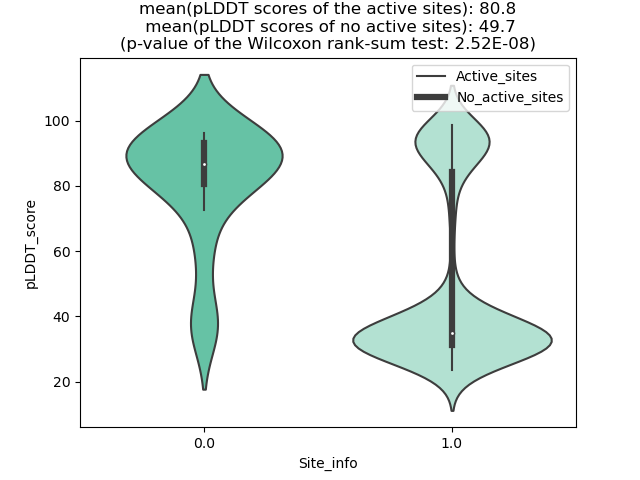
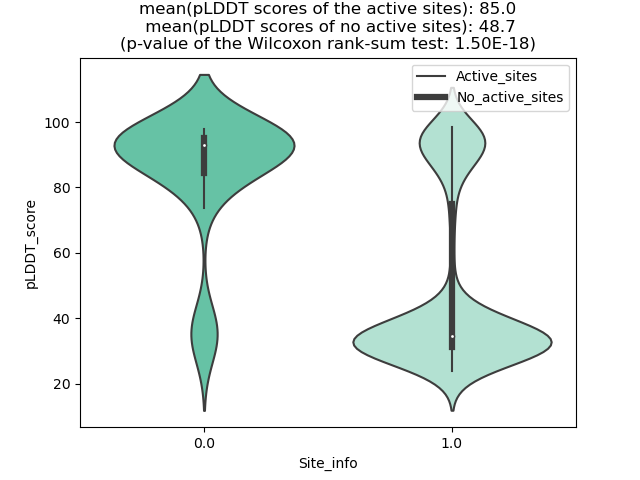
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 936 | 1.019 | 180 | 1.069 | 413.658 | 0.542 | 0.665 | 0.883 | 0.708 | 0.828 | 0.855 | 1.159 | Chain A: 36,37,38,39,40,41,43,44,45,46,47,48,49,54 ,58,62,90,92,93,94,95,96,97,98,99,123,124,125,126, 127,128,129,130 |
| 952 | 1.153 | 120 | 1.208 | 240.786 | 0.437 | 0.831 | 1.077 | 1.827 | 0.707 | 2.585 | 1.282 | Chain A: 418,422,425,426,429,430,437,440,441,446,4 50,451,452,471,472,473,1280,1281,1284 |
| 957 | 1.044 | 201 | 1.068 | 510.727 | 0.544 | 0.746 | 0.932 | 0.61 | 0.979 | 0.623 | 1.242 | Chain A: 316,317,318,319,326,329,437,440,441,444,4 45,447,448,452,453,455,456,457,461,462,463,465,466 ,467,486,488,538,539,540,541,1295,1296,1299,1300 |
| 961 | 1.032 | 268 | 1.056 | 967.946 | 0.595 | 0.732 | 0.899 | 0.547 | 0.991 | 0.552 | 1.037 | Chain A: 326,328,329,330,331,338,341,441,444,445,4 48,449,452,453,456,464,465,467,468,469,473,474,477 ,478,479,480,481,482,484,493,495,496,497,498,499,5 00,501,502,503,525,531,532,534,539,544,546,548,549 ,550,551,552,553,556,583,586,587,588,596,653,1304, 1305,1307,1308,1309,1311,1312,1315 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| RUNX1 | SUV39H1, KAT6B, CBFB, AES, PAX5, YAP1, JUN, FOS, SMAD3, SMAD1, SMAD2, SMAD5, VDR, ELF4, ELF2, CEBPB, TLE1, CBFA2T2, TRAF6, FHL2, MYOD1, HIPK2, EP300, HDAC1, HDAC3, KAT6A, tat, TCF12, TCF3, RUNX1T1, NCOR1, SIAH1, UBE2L6, RUNX1, PML, RARA, HDAC2, SIN3A, NR4A2, TRIM33, STUB1, Vdr, SMARCA4, SMARCB1, SMARCC1, CDK1, TAL1, COPRS, KMT2A, RBM14, SPEN, DNMT1, ELAVL1, CBFA2T3, PRMT1, HLA-DMA, CREBBP, UXT, CTBP2, NOTCH1, MYC, NCOR2, FOXP3, SOX2, CTBP1, HDAC11, RAG1, SPI1, ELF1, Cep72, CDK6, vif, KLF6, NFATC1, NFATC2, ZNF131, BRCA1, CDK9, FANCD2, Cbfb, CTNNB1, WWP1, RNF38, RNF44, DTX2, TRIM5, UHRF2, DZIP3, NEDD4, NKX2-1, HIF1A, ACTB, ARID1A, ARID3A, ASH2L, ACTL6A, BRD9, C2orf44, CCNK, CDC16, CDKN2AIP, CLTC, NELFB, CPSF3, CPSF6, CSTF2, CSTF2T, CTPS1, DDX1, DDX17, DDX21, DDX3X, DDX41, DDX5, DHX15, DHX36, DHX9, DPF2, EEF1A1, EEF1A2, EWSR1, EZR, FAF1, FERMT3, FIP1L1, FUBP1, FUS, GATAD2A, GATAD2B, GMPS, HIC2, HNRNPH1, HNRNPM, HNRNPR, HNRNPU, HSPA1A, HSPA2, HSPA5, HSPA8, HSPA9, IGF2BP1, INTS10, INTS9, KHSRP, KIF2B, LARP7, LCP1, LMNA, LMNB1, LRRC40, LRWD1, MED1, MED12, MED16, MED17, MED26, MEN1, MLLT1, MTA2, NDC80, NOP56, NONO, NUP85, ORC2, PABPC1, PABPC4, PAXIP1, PRMT5, PSIP1, PSPC1, RAD18, RANGAP1, RARS, RBBP5, RBM39, RPA1, RPAP2, SF1, SF3B2, SF3B3, SFPQ, SMARCC2, SMARCD1, SNRNP70, SNRPD1, SNX9, U2SURP, SRP68, SYNCRIP, TAF15, TFRC, TKT, TRAP1, TRIM25, TUBA1B, TUBB2A, UBP1, USP39, WDR76, XRCC6, XRN2, ZNF326, DMBT1, DNAJA3, DNAJB11, DPYSL2, DYNLL1, EIF3A, EIF3G, EIF4A2, EMG1, DCPS, IKZF3, IKZF1, MAD2L2, KIAA1429, Hipk2, HIST1H4A, ORF3b, MAPK1, TEAD2, SNRPC, BAG3, TEKT5, BAG4, NFKBID, VPS37C, FAM222B, OLIG3, GCC1, HGS, TEAD4, FBXO17, CIC, GSK3A, ATG7, FBXW12, RUNX2, FOXF2, NAAA, GPR55, BTF3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| RUNX1 |  |
| USP42 | 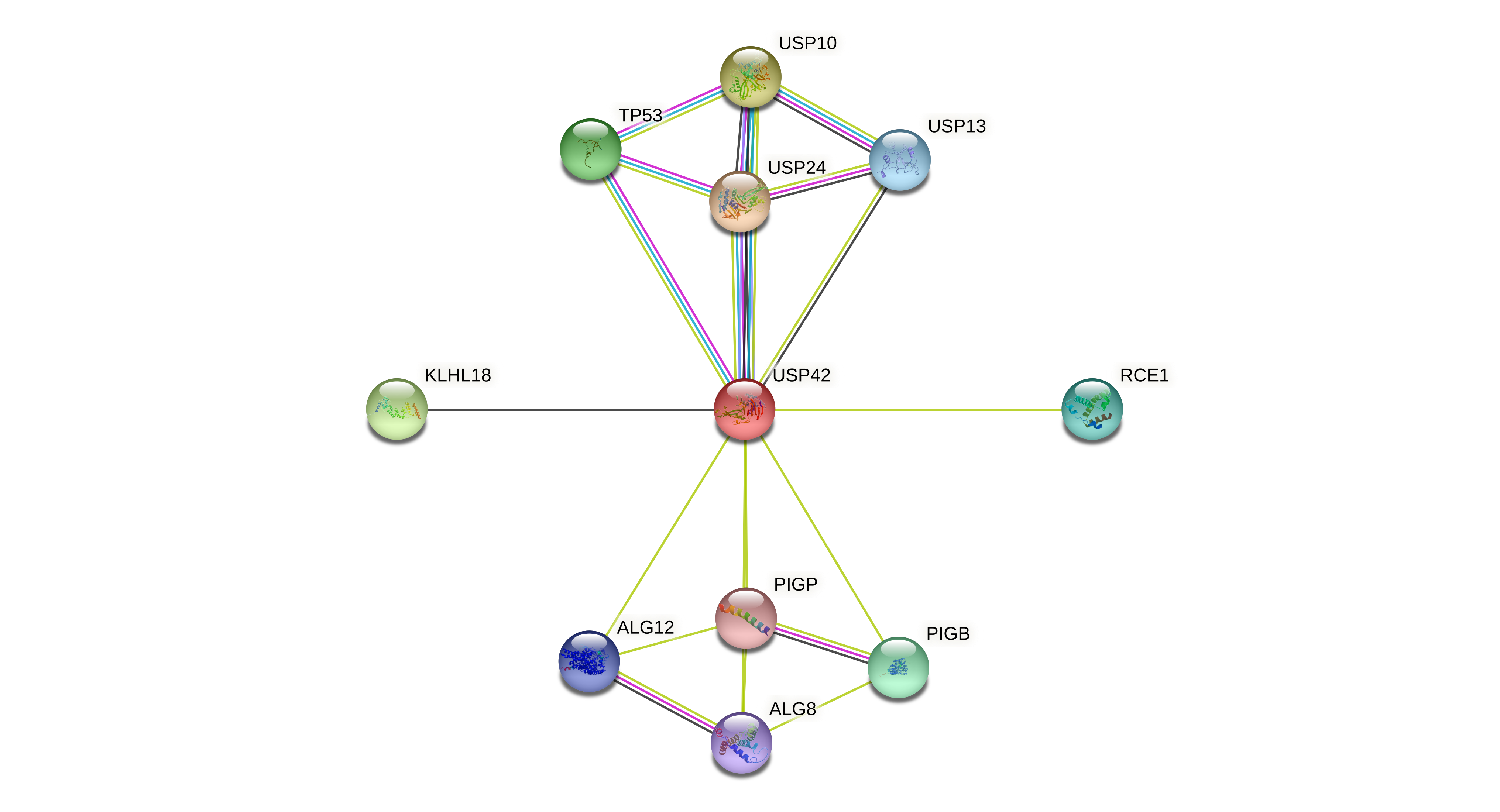 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to RUNX1-USP42 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to RUNX1-USP42 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| RUNX1 | USP42 | Lung Adenocarcinoma | MyCancerGenome | |
| RUNX1 | USP42 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| RUNX1 | USP42 | Adenocarcinoma Of The Gastroesophageal Junction | MyCancerGenome | |
| RUNX1 | USP42 | Esophageal Adenocarcinoma | MyCancerGenome | |
| RUNX1 | USP42 | Gastric Adenocarcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | RUNX1 | C1832388 | Platelet Disorder, Familial, with Associated Myeloid Malignancy | 11 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | RUNX1 | C0023467 | Leukemia, Myelocytic, Acute | 4 | CGI;CTD_human;GENOMICS_ENGLAND |
| Hgene | RUNX1 | C0026998 | Acute Myeloid Leukemia, M1 | 3 | CTD_human |
| Hgene | RUNX1 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 3 | CTD_human |
| Hgene | RUNX1 | C0023485 | Precursor B-Cell Lymphoblastic Leukemia-Lymphoma | 2 | CTD_human |
| Hgene | RUNX1 | C0003873 | Rheumatoid Arthritis | 1 | CTD_human |
| Hgene | RUNX1 | C0006413 | Burkitt Lymphoma | 1 | ORPHANET |
| Hgene | RUNX1 | C0017636 | Glioblastoma | 1 | CTD_human |
| Hgene | RUNX1 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | RUNX1 | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | RUNX1 | C0023473 | Myeloid Leukemia, Chronic | 1 | ORPHANET |
| Hgene | RUNX1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Hgene | RUNX1 | C0040034 | Thrombocytopenia | 1 | GENOMICS_ENGLAND |
| Hgene | RUNX1 | C0334588 | Giant Cell Glioblastoma | 1 | CTD_human |
| Hgene | RUNX1 | C0349639 | Juvenile Myelomonocytic Leukemia | 1 | CTD_human |
| Hgene | RUNX1 | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Hgene | RUNX1 | C1292769 | Precursor B-cell lymphoblastic leukemia | 1 | ORPHANET |
| Hgene | RUNX1 | C1621958 | Glioblastoma Multiforme | 1 | CTD_human |
| Hgene | RUNX1 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | RUNX1 | C2713368 | Hematopoetic Myelodysplasia | 1 | CTD_human |
| Hgene | RUNX1 | C3463824 | MYELODYSPLASTIC SYNDROME | 1 | CTD_human |