
|
||||||
|
Translation Factor: ERBB2 (NCBI Gene ID:2064) |
|
 Gene Summary Gene Summary |
| Gene Information | Gene Name: ERBB2 | Gene ID: 2064 | Gene Symbol | ERBB2 | Gene ID | 2064 |
| Gene Name | erb-b2 receptor tyrosine kinase 2 | |
| Synonyms | CD340|HER-2|HER-2/neu|HER2|MLN 19|NEU|NGL|TKR1 | |
| Cytomap | 17q12 | |
| Type of Gene | protein-coding | |
| Description | receptor tyrosine-protein kinase erbB-2c-erb B2/neu proteinherstatinhuman epidermal growth factor receptor 2metastatic lymph node gene 19 proteinneuro/glioblastoma derived oncogene homologneuroblastoma/glioblastoma derived oncogene homologp185erbB2 | |
| Modification date | 20200329 | |
| UniProtAcc | P04626 | |
 Child GO biological process term(s) under GO:0006412 Child GO biological process term(s) under GO:0006412 |
| GO ID | GO term |
| GO:0006417 | Regulation of translation |
| GO:0045727 | Positive regulation of translation |
| GO:0006412 | Translation |
 Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ERBB2 | GO:0007165 | signal transduction | 10572067 |
| Hgene | ERBB2 | GO:0007166 | cell surface receptor signaling pathway | 9685399 |
| Hgene | ERBB2 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 7514177 |
| Hgene | ERBB2 | GO:0014065 | phosphatidylinositol 3-kinase signaling | 7556068 |
| Hgene | ERBB2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 12000754 |
| Hgene | ERBB2 | GO:0032886 | regulation of microtubule-based process | 20937854 |
| Hgene | ERBB2 | GO:0035556 | intracellular signal transduction | 19372587 |
| Hgene | ERBB2 | GO:0042060 | wound healing | 12646923 |
| Hgene | ERBB2 | GO:0043406 | positive regulation of MAP kinase activity | 10572067 |
| Hgene | ERBB2 | GO:0045785 | positive regulation of cell adhesion | 7556068 |
| Hgene | ERBB2 | GO:0046777 | protein autophosphorylation | 7556068 |
| Hgene | ERBB2 | GO:0050679 | positive regulation of epithelial cell proliferation | 10572067 |
| Hgene | ERBB2 | GO:0071363 | cellular response to growth factor stimulus | 20010870 |
| Hgene | ERBB2 | GO:0090314 | positive regulation of protein targeting to membrane | 20010870 |
 Inferred gene age of translation factor. Inferred gene age of translation factor. |
| Gene | Inferred gene age group among (0 - 67.6], (67.6 - 355.7], (355.7 - 733], (733 - 1119.25], >1119.25 |
| ERBB2 | (733 - 1119.25] |
Top |
|
 We searched PubMed using 'ERBB2[title] AND translation [title] AND human.' We searched PubMed using 'ERBB2[title] AND translation [title] AND human.' |
| Gene | Title | PMID |
| ERBB2 | Exosome-mediated lncRNA AFAP1-AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation | 32020881 |
Top |
|
 Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. For more annotations, please visit our ExonSkipDB. |
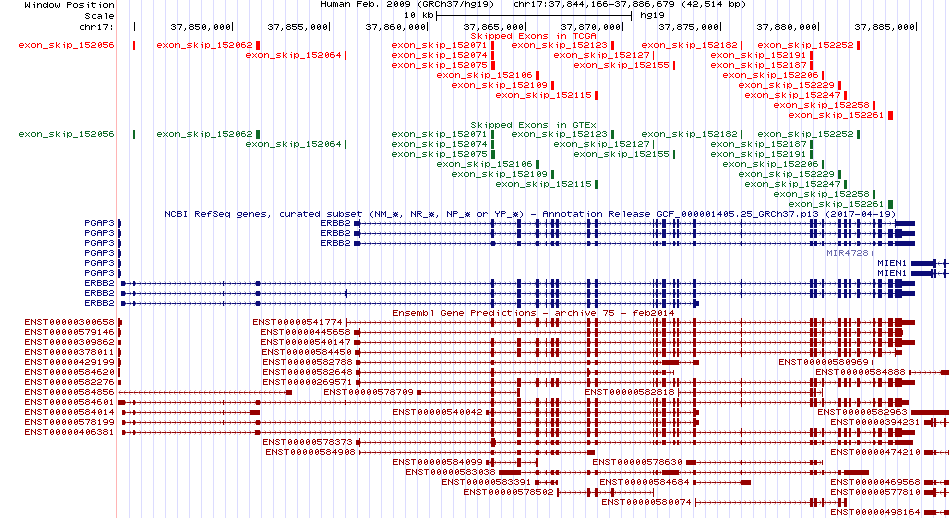 |
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ENST | Exon skip start (DNA) | Exon Skip end (DNA) | ORF |
| ENST00000269571 | 37863242 | 37863394 | Frame-shift |
| ENST00000269571 | 37865570 | 37865705 | In-frame |
| ENST00000269571 | 37866338 | 37866454 | Frame-shift |
| ENST00000269571 | 37868574 | 37868701 | Frame-shift |
| ENST00000269571 | 37871538 | 37871612 | Frame-shift |
| ENST00000269571 | 37872553 | 37872686 | Frame-shift |
| ENST00000269571 | 37876039 | 37876087 | In-frame |
| ENST00000269571 | 37879571 | 37879710 | Frame-shift |
| ENST00000269571 | 37880164 | 37880263 | In-frame |
| ENST00000269571 | 37880978 | 37881164 | In-frame |
| ENST00000269571 | 37881301 | 37881457 | In-frame |
| ENST00000269571 | 37881959 | 37882106 | In-frame |
| ENST00000269571 | 37882814 | 37882912 | Frame-shift |
| ENST00000269571 | 37883547 | 37883800 | Frame-shift |
 Exon skipping position in the amino acid sequence. Exon skipping position in the amino acid sequence. |
| ENST | Exon skip start (DNA) | Exon Skip end (DNA) | Len(transcript seq) | Exon skip start (mRNA) | Exon Skip end (mRNA) | Len(amino acid seq) | Exon skip start (AA) | Exon Skip end (AA) |
| ENST00000269571 | 37865570 | 37865705 | 4562 | 599 | 733 | 1255 | 146 | 191 |
| ENST00000269571 | 37876039 | 37876087 | 4562 | 2058 | 2105 | 1255 | 633 | 648 |
| ENST00000269571 | 37880164 | 37880263 | 4562 | 2368 | 2466 | 1255 | 736 | 769 |
| ENST00000269571 | 37880978 | 37881164 | 4562 | 2467 | 2652 | 1255 | 769 | 831 |
| ENST00000269571 | 37881301 | 37881457 | 4562 | 2653 | 2808 | 1255 | 831 | 883 |
| ENST00000269571 | 37881959 | 37882106 | 4562 | 2885 | 3031 | 1255 | 908 | 957 |
 Potentially (partially) lost protein functional features of UniProt. Potentially (partially) lost protein functional features of UniProt. |
| UniProtAcc | Exon skip start (AA) | Exon Skip end (AA) | Function feature start (AA) | Function feature end (AA) | Functional feature type | Functional feature desc. |
| P04626 | 908 | 957 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 736 | 769 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 633 | 648 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 146 | 191 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 769 | 831 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 831 | 883 | 23 | 1255 | Chain | ID=PRO_0000016669;Note=Receptor tyrosine-protein kinase erbB-2 |
| P04626 | 633 | 648 | 23 | 652 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 146 | 191 | 23 | 652 | Topological domain | Note=Extracellular;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 908 | 957 | 676 | 1255 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 736 | 769 | 676 | 1255 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 769 | 831 | 676 | 1255 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 831 | 883 | 676 | 1255 | Topological domain | Note=Cytoplasmic;Ontology_term=ECO:0000255;evidence=ECO:0000255 |
| P04626 | 908 | 957 | 720 | 987 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| P04626 | 736 | 769 | 720 | 987 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| P04626 | 769 | 831 | 720 | 987 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| P04626 | 831 | 883 | 720 | 987 | Domain | Note=Protein kinase;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| P04626 | 831 | 883 | 845 | 845 | Active site | Note=Proton acceptor;Ontology_term=ECO:0000255,ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159,ECO:0000255|PROSITE-ProRule:PRU10028 |
| P04626 | 736 | 769 | 753 | 753 | Binding site | Note=ATP;Ontology_term=ECO:0000255;evidence=ECO:0000255|PROSITE-ProRule:PRU00159 |
| P04626 | 146 | 191 | 182 | 182 | Modified residue | Note=Phosphothreonine;Ontology_term=ECO:0000244;evidence=ECO:0000244|PubMed:23186163;Dbxref=PMID:23186163 |
| P04626 | 146 | 191 | 187 | 187 | Glycosylation | Note=N-linked (GlcNAc...) asparagine;Ontology_term=ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000255;evidence=ECO:0000244|PDB:1N8Z,ECO:0000244|PDB:1S78,ECO:0000244|PDB:2A91,ECO:0000244|PDB:3N85,ECO:0000255 |
| P04626 | 146 | 191 | 162 | 192 | Disulfide bond | Ontology_term=ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244,ECO:0000244;evidence=ECO:0000244|PDB:1N8Z,ECO:0000244|PDB:1S78,ECO:0000244|PDB:2A91,ECO:0000244|PDB:3BE1,ECO:0000244|PDB:3H3B,ECO:0000244|PDB:3MZW,ECO:0000244|PDB:3N85,ECO:0000244|PDB:3WLW,ECO:0000244|PDB:3WSQ,ECO:0000244|PDB:4HRL,ECO:0000244|PDB:4HRM |
| P04626 | 633 | 648 | 626 | 634 | Disulfide bond | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3N85 |
| P04626 | 633 | 648 | 630 | 642 | Disulfide bond | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3N85 |
| P04626 | 633 | 648 | 1 | 686 | Alternative sequence | ID=VSP_039250;Note=In isoform 3. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| P04626 | 146 | 191 | 1 | 686 | Alternative sequence | ID=VSP_039250;Note=In isoform 3. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| P04626 | 146 | 191 | 1 | 610 | Alternative sequence | ID=VSP_039249;Note=In isoform 2. Missing;Ontology_term=ECO:0000305;evidence=ECO:0000305 |
| P04626 | 633 | 648 | 633 | 648 | Alternative sequence | ID=VSP_055902;Note=In isoform 6. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:24722188;Dbxref=PMID:24722188 |
| P04626 | 769 | 831 | 771 | 883 | Alternative sequence | ID=VSP_055903;Note=In isoform 6. AYVMAGVGSPYVSRLLGICLTSTVQLVTQLMPYGCLLDHVRENRGRLGSQDLLNWCMQIAKGMSYLEDVRLVHRDLAARNVLVKSPNHVKITDFGLARLLDIDETEYHADGGK->TISNLFSNFAPRGPSACCEPTCWCHSGKGQDSLPREEWGRQRRFCLWGCRGEPRVLDTPGRSCPSAPPSSCLQPSLRQPLLLGPGPTRAGGSTQHLQRDTYGREPRVPGSGRASVNQKAKSAEALMCPQGAGKA;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:24722188;Dbxref=PMID:24722188 |
| P04626 | 831 | 883 | 771 | 883 | Alternative sequence | ID=VSP_055903;Note=In isoform 6. AYVMAGVGSPYVSRLLGICLTSTVQLVTQLMPYGCLLDHVRENRGRLGSQDLLNWCMQIAKGMSYLEDVRLVHRDLAARNVLVKSPNHVKITDFGLARLLDIDETEYHADGGK->TISNLFSNFAPRGPSACCEPTCWCHSGKGQDSLPREEWGRQRRFCLWGCRGEPRVLDTPGRSCPSAPPSSCLQPSLRQPLLLGPGPTRAGGSTQHLQRDTYGREPRVPGSGRASVNQKAKSAEALMCPQGAGKA;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:24722188;Dbxref=PMID:24722188 |
| P04626 | 908 | 957 | 884 | 1255 | Alternative sequence | ID=VSP_055904;Note=In isoform 6. Missing;Ontology_term=ECO:0000303;evidence=ECO:0000303|PubMed:24722188;Dbxref=PMID:24722188 |
| P04626 | 736 | 769 | 755 | 755 | Natural variant | ID=VAR_055432;Note=In LNCR%3B somatic mutation%3B unknown pathological significance. L->P;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15457249;Dbxref=dbSNP:rs121913469,PMID:15457249 |
| P04626 | 736 | 769 | 768 | 768 | Natural variant | ID=VAR_042097;Note=L->S;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:17344846;Dbxref=dbSNP:rs56366519,PMID:17344846 |
| P04626 | 769 | 831 | 774 | 774 | Natural variant | ID=VAR_055433;Note=In LNCR%3B somatic mutation%3B unknown pathological significance. M->MAYVM;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15457249;Dbxref=PMID:15457249 |
| P04626 | 769 | 831 | 776 | 776 | Natural variant | ID=VAR_042098;Note=In GASC%3B somatic mutation%3B unknown pathological significance. G->S;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:15457249,ECO:0000269|PubMed:17344846;Dbxref=dbSNP:rs28933369,PMID:15457249,PMID:17344846 |
| P04626 | 769 | 831 | 779 | 779 | Natural variant | ID=VAR_055434;Note=In LNCR%3B somatic mutation%3B unknown pathological significance. S->SVGS;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15457249;Dbxref=PMID:15457249 |
| P04626 | 831 | 883 | 857 | 857 | Natural variant | ID=VAR_042099;Note=In OC%3B somatic mutation%3B unknown pathological significance. N->S;Ontology_term=ECO:0000269,ECO:0000269;evidence=ECO:0000269|PubMed:15457249,ECO:0000269|PubMed:17344846;Dbxref=dbSNP:rs28933370,PMID:15457249,PMID:17344846 |
| P04626 | 908 | 957 | 914 | 914 | Natural variant | ID=VAR_055435;Note=In GLM%3B somatic mutation%3B unknown pathological significance. E->K;Ontology_term=ECO:0000269;evidence=ECO:0000269|PubMed:15457249;Dbxref=dbSNP:rs28933368,PMID:15457249 |
| P04626 | 146 | 191 | 147 | 150 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:2A91 |
| P04626 | 146 | 191 | 152 | 156 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5MY6 |
| P04626 | 146 | 191 | 164 | 166 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3H3B |
| P04626 | 146 | 191 | 169 | 172 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:5MY6 |
| P04626 | 146 | 191 | 175 | 177 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:1N8Z |
| P04626 | 146 | 191 | 182 | 184 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3H3B |
| P04626 | 633 | 648 | 635 | 637 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3N85 |
| P04626 | 736 | 769 | 730 | 739 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 736 | 769 | 748 | 755 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 736 | 769 | 761 | 774 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 769 | 831 | 761 | 774 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 769 | 831 | 785 | 799 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 769 | 831 | 806 | 812 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 769 | 831 | 814 | 816 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 769 | 831 | 819 | 838 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 831 | 883 | 819 | 838 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 831 | 883 | 848 | 850 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 831 | 883 | 851 | 855 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 831 | 883 | 858 | 861 | Beta strand | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 908 | 957 | 901 | 916 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 908 | 957 | 922 | 925 | Turn | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 908 | 957 | 928 | 930 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 908 | 957 | 931 | 936 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
| P04626 | 908 | 957 | 949 | 958 | Helix | Ontology_term=ECO:0000244;evidence=ECO:0000244|PDB:3PP0 |
Top |
|
 Gene expression level across TCGA pancancer Gene expression level across TCGA pancancer |
 |
 Gene expression level across GTEx pantissue Gene expression level across GTEx pantissue |
 |
 Expression level of gene isoforms across TCGA pancancer Expression level of gene isoforms across TCGA pancancer |
 |
 Expression level of gene isoforms across GTEx pantissue Expression level of gene isoforms across GTEx pantissue |
 |
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution Cancer(tissue) type-specific expression level of Translation factor using z-score distriution |
 |
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples) Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples) |
 |
| Cancer type | Translation factor | FC | adj.pval |
| STAD | ERBB2 | 2.08308782318447 | 0.00733334058895707 |
| BLCA | ERBB2 | 1.70986293820428 | 0.014068603515625 |
| BRCA | ERBB2 | 2.10379074903324 | 4.64802044953281e-07 |
| THCA | ERBB2 | 1.02793927266444 | 5.71717197842929e-07 |
| KICH | ERBB2 | -2.13618034410539 | 6.55651092529297e-06 |
| LUAD | ERBB2 | 1.65046120667728 | 8.94245335020896e-05 |
| HNSC | ERBB2 | -3.64938375773393 | 9.1349647846073e-09 |
Top |
|
 Translation factor expression regulation through miRNA binding Translation factor expression regulation through miRNA binding |
| Cancer type | Gene | miRNA | TargetScan binding score (Context++ score percentile) | Coefficient | Pvalue |
 Translation factor expression regulation through methylation in the promoter of Translation factor Translation factor expression regulation through methylation in the promoter of Translation factor |
 |
| Cancer type | Gene | methyl group b | methyl group a | DEG pval | avg methyl in b | avg methyl in a | avg exp in b | avg exp in a |
| LIHC | ERBB2 | 2 | 1 | 0.00856161870864884 | 0.24412712890625 | 0.176576184971098 | 0.158119715268625 | 0.0270168047737106 |
| TGCT | ERBB2 | 2 | 1 | 0.0452907458711372 | 0.213704953560371 | 0.156055736463461 | 0.669519048907529 | 0.358306903330417 |
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation) |
 |
| Cancer type | Gene | methyl group b | methyl group a | DEG pval | avg methyl in b | avg methyl in a | avg exp in b | avg exp in a |
 Translation factor expression regulation through copy number variation of Translation factor Translation factor expression regulation through copy number variation of Translation factor |
 |
| Cancer type | Gene | Coefficient | Pvalue |
| LGG | ERBB2 | -0.060290344 | 0.049858555 |
Top |
|
 Strongly correlated genes belong to cellular important gene groups with ERBB2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green) Strongly correlated genes belong to cellular important gene groups with ERBB2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green) |
 |
| Cancer type | Gene group | Translation factor | Correlated gene | Coefficient | Pvalue |
| TGCT | Cell metabolism gene | ERBB2 | AGRN | 0.813274397 | 4.79E-38 |
| TGCT | Cell metabolism gene | ERBB2 | GPC6 | 0.820558741 | 3.02E-39 |
| TGCT | Cell metabolism gene | ERBB2 | SLC44A3 | 0.823648715 | 9.00E-40 |
| TGCT | Cell metabolism gene | ERBB2 | SULT1E1 | 0.831064207 | 4.46E-41 |
| TGCT | Cell metabolism gene | ERBB2 | TEAD3 | 0.863399174 | 1.30E-47 |
| TGCT | CGC | ERBB2 | BMP5 | 0.810556345 | 1.30E-37 |
| TGCT | CGC | ERBB2 | PTPRK | 0.832837208 | 2.13E-41 |
| TGCT | CGC | ERBB2 | TMPRSS2 | 0.870838832 | 2.35E-49 |
| TGCT | Epifactor | ERBB2 | FOXP4 | 0.818966676 | 5.58E-39 |
| TGCT | IUPHAR | ERBB2 | SLC15A1 | 0.802855792 | 2.04E-36 |
| TGCT | IUPHAR | ERBB2 | CFTR | 0.806333555 | 5.97E-37 |
| TGCT | IUPHAR | ERBB2 | LGR5 | 0.810655011 | 1.26E-37 |
| TGCT | IUPHAR | ERBB2 | ABCC6 | 0.816116297 | 1.65E-38 |
| TGCT | IUPHAR | ERBB2 | ITGB6 | 0.816420545 | 1.47E-38 |
| TGCT | IUPHAR | ERBB2 | HSD11B2 | 0.819662358 | 4.27E-39 |
| TGCT | IUPHAR | ERBB2 | SLC44A3 | 0.823648715 | 9.00E-40 |
| TGCT | IUPHAR | ERBB2 | PTPRK | 0.832837208 | 2.13E-41 |
| TGCT | IUPHAR | ERBB2 | ADAMTS7 | 0.833429014 | 1.66E-41 |
| TGCT | IUPHAR | ERBB2 | HNF4G | 0.852223304 | 3.53E-45 |
| TGCT | IUPHAR | ERBB2 | CRABP2 | 0.863212273 | 1.43E-47 |
| TGCT | IUPHAR | ERBB2 | TMPRSS2 | 0.870838832 | 2.35E-49 |
| TGCT | TF | ERBB2 | SOX11 | 0.801256613 | 3.55E-36 |
| TGCT | TF | ERBB2 | FOXP4 | 0.818966676 | 5.58E-39 |
| TGCT | TF | ERBB2 | HIF3A | 0.82099774 | 2.55E-39 |
| TGCT | TF | ERBB2 | HES1 | 0.821910057 | 1.78E-39 |
| TGCT | TF | ERBB2 | SPDEF | 0.822165677 | 1.61E-39 |
| TGCT | TF | ERBB2 | HNF4G | 0.852223304 | 3.53E-45 |
| TGCT | TF | ERBB2 | TEAD3 | 0.863399174 | 1.30E-47 |
| TGCT | TSG | ERBB2 | SOX11 | 0.801256613 | 3.55E-36 |
| TGCT | TSG | ERBB2 | CFTR | 0.806333555 | 5.97E-37 |
| TGCT | TSG | ERBB2 | VIL1 | 0.811601258 | 8.88E-38 |
| TGCT | TSG | ERBB2 | BMP4 | 0.826475636 | 2.91E-40 |
| TGCT | TSG | ERBB2 | SEMA3B | 0.831127836 | 4.34E-41 |
| TGCT | TSG | ERBB2 | PTPRK | 0.832837208 | 2.13E-41 |
| TGCT | TSG | ERBB2 | KRT19 | 0.878800781 | 2.40E-51 |
| THYM | IUPHAR | ERBB2 | ITGB8 | 0.814042882 | 4.29E-30 |
| UCS | IUPHAR | ERBB2 | ITGB8 | 0.814042882 | 4.29E-30 |
| UVM | Cell metabolism gene | ERBB2 | SMG6 | 0.833245116 | 9.08E-22 |
| UVM | Cell metabolism gene | ERBB2 | MED24 | 0.848376593 | 3.01E-23 |
| UVM | CGC | ERBB2 | DCTN1 | 0.832665428 | 1.03E-21 |
| UVM | CGC | ERBB2 | LASP1 | 0.858367578 | 2.57E-24 |
Top |
|
 Protein 3D structure Protein 3D structureVisit iCn3D. |
Top |
|
 Protein-protein interaction networks Protein-protein interaction networks * Overlap between up-regulated DEGs (log2FC<-1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) |
 |
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P)) |
 |
 * Edge colors based on TCGA cancer types. |
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P)) * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P)) |
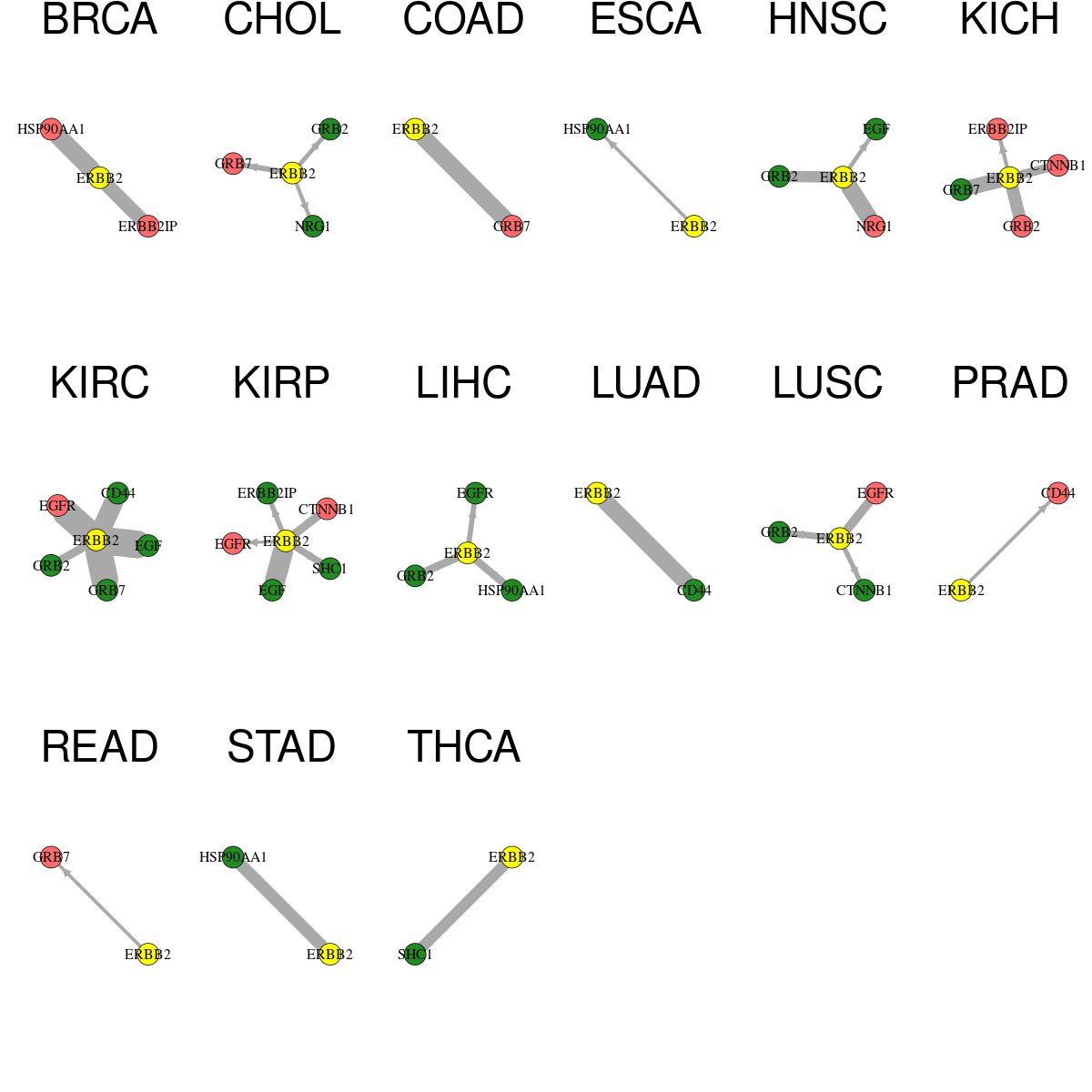 |
| Cancer type | Translation factor | Interacting protein coding gene | FC | adj.pval |
| LUSC | ERBB2 | EGFR | 1.32757138391417 | 0.000102202753672845 |
| KICH | ERBB2 | CTNNB1 | 1.35387002426383 | 0.000139892101287842 |
| KIRP | ERBB2 | CTNNB1 | 1.44413647030576 | 0.00019507110118866 |
| KIRC | ERBB2 | GRB2 | -3.50962289529079 | 0.000210123708729258 |
| LIHC | ERBB2 | GRB2 | -2.6070669266439 | 0.000329386776931513 |
| LIHC | ERBB2 | HSP90AA1 | -1.87137739671902 | 0.0003817245773207 |
| KIRP | ERBB2 | SHC1 | -1.28530866485745 | 0.000906126573681831 |
| LUSC | ERBB2 | GRB2 | -1.60175668402919 | 0.00116257561090457 |
| CHOL | ERBB2 | GRB7 | 2.57684563734882 | 0.00390625 |
| LIHC | ERBB2 | EGFR | -1.2230538094017 | 0.00496795806084514 |
| KIRP | ERBB2 | ERBB2IP | -1.61124702030031 | 0.00608485564589501 |
| LUSC | ERBB2 | CTNNB1 | -1.53686315879634 | 0.00765883184752251 |
| CHOL | ERBB2 | GRB2 | -1.28858368343854 | 0.01171875 |
| HNSC | ERBB2 | EGF | -3.20486700850326 | 0.0149245594448075 |
| KICH | ERBB2 | ERBB2IP | 1.12033403670127 | 0.0173123478889465 |
| CHOL | ERBB2 | NRG1 | -3.34852291534279 | 0.01953125 |
| PRAD | ERBB2 | CD44 | 1.04545382452033 | 0.0305494319135424 |
| READ | ERBB2 | GRB7 | 4.00121793802205 | 0.03125 |
| ESCA | ERBB2 | HSP90AA1 | -2.95407832928575 | 0.0322265625 |
| KIRP | ERBB2 | EGFR | 1.14056399927375 | 0.0394268441013992 |
| HNSC | ERBB2 | GRB2 | -2.71037834855917 | 1.00141839993739e-05 |
| BRCA | ERBB2 | HSP90AA1 | 2.1978220577015 | 1.13731762084626e-08 |
| THCA | ERBB2 | SHC1 | -1.92572983257134 | 1.13787415943499e-05 |
| KIRC | ERBB2 | GRB7 | -2.08557276759602 | 1.28432097821042e-12 |
| KICH | ERBB2 | GRB7 | -1.35886098374196 | 1.78813934326172e-07 |
| KIRC | ERBB2 | EGFR | 1.55836383937799 | 2.25998111659284e-12 |
| KICH | ERBB2 | GRB2 | 1.69470901099239 | 2.5629997253418e-06 |
| HNSC | ERBB2 | NRG1 | 1.15540013951039 | 2.86950125882868e-08 |
| COAD | ERBB2 | GRB7 | 1.93627228443447 | 2.98023223876954e-08 |
| STAD | ERBB2 | HSP90AA1 | -1.65705125430285 | 3.51201742887497e-06 |
| LUAD | ERBB2 | CD44 | -1.69965776529045 | 3.53668114331368e-08 |
| KIRC | ERBB2 | CD44 | -1.14543500743668 | 3.61971983207531e-09 |
| BRCA | ERBB2 | ERBB2IP | 1.50051933270829 | 4.79363542815941e-07 |
| KIRC | ERBB2 | EGF | -2.11944460092247 | 6.15531645908303e-13 |
| KIRP | ERBB2 | EGF | -2.43867553870574 | 9.31322574615479e-10 |
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Protein-protein interactors with this translation factor (BIOGRID-3.4.160) |
| PPI interactors with ERBB2 |
| SHC1, SH3BGRL, PTPN11, GRB2, MUC1, CTNNB1, ERBB2, CD82, EGF, PAK1, ERBB2IP, PICK1, PIK3R2, DLG4, ERBB4, Errfi1, NRG1, JUP, STAT3, SRC, JAK2, HSP90B1, CD44, GRB7, PLCG1, IL6ST, OSMR, PTK2B, HSP90AA1, ELF3, EGFR, ERBB3, HDAC6, LRIG1, STUB1, HSPA4, HSP82, IGF1R, CUL5, ESR1, PPP1R13B, H2AFY, PIK3R1, PIN1, PTK6, ACTB, POLR1B, KPNB1, XPO1, CLTC, AP2B1, EEA1, APBB1, SH2B2, IRS4, SH2D1B, SOCS1, NCK2, DAB1, GRAP2, APBB3, ABL1, FGR, CHN1, FER, PLCG2, RASA1, JAK1, IRS1, STAT1, MATK, TEC, TXK, ABL2, SYK, CRK, CRKL, BLK, BMX, VAV2, SH3BP2, SHC2, ITK, SLA, MAPK8IP2, RIN1, BCAR3, TENC1, TNS3, DOK6, SH2D5, SUPT6H, ANKS1B, CLNK, DOK4, BLNK, RIN2, SHC3, PIK3R3, ANKS1A, VAV1, DOK1, SH2D3A, SLA2, SH2D2A, CISH, VAV3, MAPK8IP1, SH2B3, TLN1, SRPK1, CBL, ERRFI1, FRS3, USP2, NXF1, DTWD2, TGFBR2, NAALADL2, LPAR6, VSIG4, PTGER3, CYP17A1, GLCE, IL13RA2, KLK5, LYPD3, NAT2, PRDM14, PSMC3IP, RHOBTB2, THRSP, ANXA6, BAI1, CLCN6, GMPR2, HDLBP, IARS, MTCH1, MUCL1, PHF23, POLD2, PSAP, STK24, TCEAL2, ATP6AP1, TTC9B, UTP18, ATP1A1, EEF1G, TNPO1, PC, SURF4, TFRC, VAPB, PTDSS1, HPSE, ZNF280C, CCDC88A, ZNF622, Tor1aip1, BTC, PTK2, TGFA, BCAR1, ESRRB, CDC37, EZR, RDX, ITGAV, L1CAM, ITGB1, CHEK2, TOP1, RRM2, CDK4, CDH2, CDH1, PTPN6, PTPRR, PTPRT, PTPRU, PTPRA, PTPRH, MPZL2, SSH1, CD79B, MAS1, HAVCR2, IL4R, PPM1A, PPM1F, ILKAP, DUSP14, DUSP18, DUSP19, STYX, GAB1, HNRNPL, OPCML, CPNE3, FBXO7, HSP90AB1, PLA2G2A, CXCL8, DOCK7, ADRB2, BECN1, SDC1, MUC4, CDC25A, PRKDC, CAMK2G, CDK12, GPX1, FGFR3, TK1, PIK3CB, BIRC5, PIK3CA, ADRBK1, GRIK5, CALM1, LRIG3, MCL1, KIAA1429, PDGFRB, EPHA1, EPHA2, EPHA3, EPHA4, EPHA5, EPHB1, EPHB2, EPHB3, EPHB6, FGFR1, FGFR2, FGFR4, INSR, INSRR, MST1R, MET, PDGFRA, PTK7, RET, ROR2, NTRK1, NTRK2, NTRK3, FLT3, FLT4, STYK1, ALK, MERTK, TYRO3, DDR1, TEK, CSK, HRAS, APEX1, PSMD3, CCND2, CDKN2B, LATS2, STK11, FZR1, GLIS2, MAP2K5, TTYH2, ABCB5, ARNT, CBLC, AKT1, AURKA, CDK6, CDKN2C, GRM1, HGF, KDELR2, MAP2K3, MYC, NF2, RAF1, RASSF1, TP53, ORF7b, AATK, ACP1, AIP, ALDOA, AP3B2, APLP1, ARF1, ARF5, ASRGL1, ATP6V0C, BNIP1, C10orf35, C14orf1, TMEM230, C21orf59, SMIM20, CBR1, CEND1, CLASP2, COL9A2, COL9A3, CRIP2, DST, FKBP3, FKBP8, FLNB, GABARAPL1, GABARAPL2, GLRX3, GPM6B, GAPDH, IRF2BP2, ITPKA, ITPR1, JMJD4, KCTD6, KLC1, LGALS8, LPCAT4, LRRC7, MDH1, MSN, MYO9A, NEFH, NINL, NUDT16L1, ODF2L, PDCD11, PGAM1, PIN4, POU6F1, PPA1, PSMA1, PTGES3L-AARSD1, RBM15B, RHBDD2, RPRML, RTN4, S100A11, SAFB2, SELK, SLC7A4, SMPD4, SNAP91, SPP1, STMN2, SYNM, TMEM134, TSEN15, TXN, TYMP, ZFAND1, ZMAT2, ZMYND8, ZNF532, RAB5C, TMA7, ILF3, AKAP12, HSPA9, HSPA1A, CCT3, PABPC1, YWHAZ, HSPD1, HSPA5, VAPA, STIP1, PSMC3, MEPCE, TRIP6, CACYBP, FKBP1A, STMN1, ALDOC, TVP23B, CSDE1, GSN, CFL1, PRDX5, CKB, PEBP1, nsp4, ORF3a, ORF7a, S, M, BAAT, C9orf156, CDK8, CYLC2, FBP1, FRAT2, HSD17B3, CCDC180, MAP2K1, SFRP4, SMAD1, TGFB1, ZNF189, TRIM66, FES, GRB10, GRB14, HCK, HSH2D, INPPL1, LCK, LCP2, LYN, NCK1, SH2B1, SH2D1A, SH2D3C, SHB, SHC4, SHD, SOCS3, SOCS5, SOCS6, STAP1, TNS1, YES1, ZAP70, HAX1, E5b, CUL4A, MKRN2, C11orf52, CAV1, FLOT1, GJA1, KRAS, OCLN, RAB35, STX6, AIM1, TIMP1, SLITRK3, SPCS1, CX3CL1, HCST, TMEM200A, ARL15, TICAM2, IGDCC4, ARL4D, PIGH, RELT, OPALIN, AQP3, ACVR1, AMHR2, EFNB1, SORL1, FBXW7, DEPTOR, BTRC, |
Top |
|
 Clinically associated variants from ClinVar. Clinically associated variants from ClinVar. |
| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| ERBB2 | chr17 | 37855834 | C | A | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37856534 | C | T | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863238 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863250 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001623|5_prime_UTR_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863262 | G | T | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001582|initiatior_codon_variant,SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863268 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863280 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863302 | A | G | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37863309 | G | A | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864569 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864584 | A | C | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864606 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864646 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864655 | C | T | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001587|nonsense,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864691 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864693 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864713 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864729 | C | G | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864731 | C | G | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864776 | G | A | single_nucleotide_variant | Benign/Likely_benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37864781 | C | T | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37865601 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37865629 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37865659 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37865694 | G | A | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866082 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866084 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866132 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866343 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866370 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866383 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866412 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866429 | C | T | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1|not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866430 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866583 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866588 | C | T | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866601 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866641 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866649 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866689 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866707 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37866741 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37868186 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37868205 | G | A | single_nucleotide_variant | Likely_pathogenic | Lung_adenocarcinoma | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37868205 | G | C | single_nucleotide_variant | Likely_pathogenic | Breast_neoplasm | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37868208 | C | A | single_nucleotide_variant | Pathogenic | Neoplasm|Squamous_cell_carcinoma_of_the_skin|Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Lung_adenocarcinoma|Neoplasm_of_uterine_cervix|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Ovarian_Serous_Cystadenocarcinoma|Squamous_cel | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37868208 | C | T | single_nucleotide_variant | Likely_pathogenic | Neoplasm|Squamous_cell_carcinoma_of_the_skin|Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Lung_adenocarcinoma|Neoplasm_of_uterine_cervix|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Ovarian_Serous_Cystadenocarcinoma|Squamous_cel | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37868319 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37868331 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37868348 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37868618 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant |
| ERBB2 | chr17 | 37868691 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37871547 | C | A | single_nucleotide_variant | Likely_benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37871559 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant |
| ERBB2 | chr17 | 37871707 | T | G | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37871724 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37871745 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37871770 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37871996 | C | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37871996 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872005 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872035 | G | T | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872045 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872050 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872064 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872068 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872095 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872145 | C | T | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872146 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872168 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872174 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872558 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872607 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872609 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872646 | CG | C | Deletion | Pathogenic | Malignant_Colorectal_Neoplasm | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872647 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872696 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872780 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872834 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37872846 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873566 | C | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873568 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Glioma_susceptibility_1|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873581 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873607 | A | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873628 | C | A | single_nucleotide_variant | Uncertain_significance | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873698 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37873743 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant | SO:0001624|3_prime_UTR_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37876073 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879564 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879585 | A | G | single_nucleotide_variant | Benign | ERBB2_POLYMORPHISM|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879588 | A | G | single_nucleotide_variant | Benign | ERBB2_POLYMORPHISM|not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879615 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879658 | G | A | single_nucleotide_variant | Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Adenocarcinoma_of_prostate|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879673 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879834 | C | T | single_nucleotide_variant | Pathogenic | VISCERAL_NEUROPATHY,_FAMILIAL,_2,_AUTOSOMAL_RECESSIVE | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879853 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37879877 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880218 | GTTGAGGGAAAACACA | G | Deletion | Pathogenic | Breast_neoplasm | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001822|inframe_deletion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880219 | T | A | single_nucleotide_variant | Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Malignant_melanoma_of_skin|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Papillary_renal_cell_carcinoma,_sporadic|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880219 | TT | CC | Indel | Pathogenic/Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Malignant_melanoma_of_skin|Lung_adenocarcinoma|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Papillary_renal_cell_carcinoma,_sporadic|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880220 | T | C | single_nucleotide_variant | Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Malignant_melanoma_of_skin|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Papillary_renal_cell_carcinoma,_sporadic|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880220 | T | G | single_nucleotide_variant | Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Malignant_melanoma_of_skin|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Papillary_renal_cell_carcinoma,_sporadic|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880261 | G | A | single_nucleotide_variant | Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Carcinoma_of_esophagus|Neoplasm_of_uterine_cervix|Adenocarcinoma_of_stomach | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880261 | G | C | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Carcinoma_of_esophagus|Neoplasm_of_uterine_cervix|Adenocarcinoma_of_stomach | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880261 | G | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Transitional_cell_carcinoma_of_the_bladder|Breast_neoplasm|Carcinoma_of_esophagus|Neoplasm_of_uterine_cervix|Adenocarcinoma_of_stomach | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880270 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880981 | A | AGCATACGTGATG | Duplication | Likely_pathogenic | Lung_adenocarcinoma|Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880984 | A | ATACGTGATGGCT | Duplication | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880986 | A | ACGTGATGGCCTC | Insertion | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880986 | A | ACGTGATGGCTTC | Insertion | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880988 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880990 | GA | G | Deletion | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880997 | G | A | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Neoplasm|Gastric_cancer|Adenocarcinoma_of_stomach | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880997 | G | GTCT | Insertion | Uncertain_significance | not_specified | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel |
| ERBB2 | chr17 | 37880997 | G | GTGT | Insertion | Likely_pathogenic | Breast_neoplasm|Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel |
| ERBB2 | chr17 | 37880997 | G | GTTT | Insertion | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel |
| ERBB2 | chr17 | 37880997 | G | TTAT | Indel | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel | SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant,SO:0001820|inframe_indel |
| ERBB2 | chr17 | 37880998 | G | GTGTGGGCTC | Duplication | Pathogenic | Lung_adenocarcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37880998 | G | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881000 | G | A | single_nucleotide_variant | Likely_pathogenic | Breast_neoplasm|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881000 | G | C | single_nucleotide_variant | Likely_pathogenic | Breast_neoplasm | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881000 | G | T | single_nucleotide_variant | Uncertain_significance | Breast_neoplasm|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881001 | T | TGGGCTCCCC | Duplication | Pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881002 | G | GGGCTCCCCA | Duplication | Likely_pathogenic | Non-small_cell_lung_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881003 | G | GGGCTCCCCA | Insertion | Pathogenic | Breast_neoplasm | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001821|inframe_insertion,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881025 | T | C | single_nucleotide_variant | Likely_pathogenic | Breast_neoplasm | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881050 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881071 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881102 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881111 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881114 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881143 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881169 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881299 | C | T | single_nucleotide_variant | not_provided | Familial_cancer_of_breast | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881314 | C | T | single_nucleotide_variant | not_provided | Endometrial_carcinoma | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881328 | G | A | single_nucleotide_variant | not_provided | Familial_cancer_of_breast | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881332 | G | A | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Pancreatic_adenocarcinoma|Breast_neoplasm|Adenocarcinoma_of_stomach|Neoplasm_of_the_large_intestine|Carcinoma_of_gallbladder|Uterine_Carcinosarcoma|Malignant_neoplasm_of_body_of_uterus | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881343 | C | T | single_nucleotide_variant | not_provided | Familial_cancer_of_breast | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881344 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881349 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881376 | C | T | single_nucleotide_variant | not_provided | Familial_cancer_of_breast | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881378 | A | G | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Neoplasm_of_ovary|Ovarian_Adenocarcinoma | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881413 | C | T | single_nucleotide_variant | not_provided | Neoplasm_of_ovary | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881414 | T | G | single_nucleotide_variant | Pathogenic | Lung_adenocarcinoma | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881575 | C | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881616 | C | T | single_nucleotide_variant | Pathogenic | Breast_neoplasm | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881618 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881636 | G | C | single_nucleotide_variant | Uncertain_significance | Malignant_tumor_of_prostate | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881654 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37881974 | G | A | single_nucleotide_variant | Pathogenic | Glioma_susceptibility_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882003 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882020 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882024 | G | T | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882060 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882110 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37882856 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883113 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883133 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883141 | G | A | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883167 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883175 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883178 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883183 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883205 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883212 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883243 | G | A | single_nucleotide_variant | not_provided | Familial_cancer_of_breast | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883246 | C | T | single_nucleotide_variant | Uncertain_significance | Glioma_susceptibility_1 | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883253 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883257 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001575|splice_donor_variant,SO:0001627|intron_variant | SO:0001575|splice_donor_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883540 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883570 | T | C | single_nucleotide_variant | Uncertain_significance | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883638 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883656 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883696 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883720 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883748 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883763 | T | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883784 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883791 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001627|intron_variant |
| ERBB2 | chr17 | 37883956 | C | A | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883957 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883959 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883965 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883966 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883970 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37883982 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884011 | G | A | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884013 | C | CG | Insertion | Pathogenic | Malignant_Colorectal_Neoplasm | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001589|frameshift_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884013 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884037 | C | G | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884046 | C | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884050 | C | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884069 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884078 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884120 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884149 | C | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884159 | T | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884176 | C | A | single_nucleotide_variant | Benign | not_specified|not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884180 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884191 | A | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884218 | G | T | single_nucleotide_variant | not_provided | not_specified | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884234 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884270 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant | SO:0001619|non-coding_transcript_variant,SO:0001819|synonymous_variant,SO:0001624|3_prime_UTR_variant |
| ERBB2 | chr17 | 37884292 | G | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant | SO:0001583|missense_variant,SO:0001619|non-coding_transcript_variant,SO:0001624|3_prime_UTR_variant |
 nsSNVs with sample frequency (size of circle) from TCGA 33 cancers. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers. |
 SNVs and Indels SNVs and Indels |
| Gene | Cancer type | Chromosome | Start | End | RefSeeq | MutSeq | Mutation type | AAchange | # samples |
| ERBB2 | BLCA | chr17 | 37868208 | 37868208 | C | T | Missense_Mutation | 39 | |
| ERBB2 | BLCA | chr17 | 37868208 | 37868208 | C | T | Missense_Mutation | p.S280F | 31 |
| ERBB2 | BLCA | chr17 | 37868208 | 37868208 | C | T | Missense_Mutation | p.S310F | 18 |
| ERBB2 | BLCA | chr17 | 37880220 | 37880220 | T | C | Missense_Mutation | p.L755S | 16 |
| ERBB2 | BRCA | chr17 | 37881332 | 37881332 | G | A | Missense_Mutation | p.V842I | 14 |
| ERBB2 | BRCA | chr17 | 37879658 | 37879658 | G | A | Missense_Mutation | p.R678Q | 10 |
| ERBB2 | BLCA | chr17 | 37881000 | 37881000 | G | T | Missense_Mutation | 8 | |
| ERBB2 | LUAD | chr17 | 37880981 | 37880982 | - | GCATACGTGATG | In_Frame_Ins | p.740_740D>EHT*C | 8 |
| ERBB2 | BLCA | chr17 | 37881332 | 37881332 | G | A | Missense_Mutation | p.V812I | 7 |
| ERBB2 | BRCA | chr17 | 37881000 | 37881000 | G | C | Missense_Mutation | p.V777L | 7 |
| ERBB2 | BLCA | chr17 | 37881332 | 37881332 | G | A | Missense_Mutation | 7 | |
| ERBB2 | LUAD | chr17 | 37864639 | 37864639 | G | A | Silent | p.Q67Q | 6 |
| ERBB2 | BRCA | chr17 | 37880261 | 37880261 | G | T | Missense_Mutation | p.D769Y | 6 |
| ERBB2 | CHOL | chr17 | 37880219 | 37880219 | T | C | Silent | p.L755L | 5 |
| ERBB2 | BLCA | chr17 | 37879658 | 37879658 | G | A | Missense_Mutation | p.R648Q | 5 |
| ERBB2 | BLCA | chr17 | 37881000 | 37881000 | G | T | Missense_Mutation | p.V747L | 5 |
| ERBB2 | BRCA | chr17 | 37880261 | 37880261 | G | C | Missense_Mutation | p.D769H | 4 |
| ERBB2 | ESCA | chr17 | 37881000 | 37881000 | G | A | Missense_Mutation | p.V777M | 4 |
| ERBB2 | BLCA | chr17 | 37868208 | 37868208 | C | A | Missense_Mutation | p.S280Y | 4 |
| ERBB2 | LIHC | chr17 | 37882888 | 37882888 | C | - | Frame_Shift_Del | p.D952fs | 4 |
| ERBB2 | BLCA | chr17 | 37880220 | 37880220 | T | C | Missense_Mutation | 4 | |
| ERBB2 | BLCA | chr17 | 37880261 | 37880261 | G | A | Missense_Mutation | 4 | |
| ERBB2 | UCEC | chr17 | 37879613 | 37879613 | T | C | Missense_Mutation | p.L663P | 3 |
| ERBB2 | HNSC | chr17 | 37881982 | 37881982 | G | A | Missense_Mutation | p.M886I | 3 |
| ERBB2 | UCEC | chr17 | 37883626 | 37883626 | G | T | Missense_Mutation | p.G1080W | 3 |
| ERBB2 | BLCA | chr17 | 37880220 | 37880220 | T | C | Missense_Mutation | p.L725S | 3 |
| ERBB2 | BLCA | chr17 | 37881392 | 37881392 | A | G | Missense_Mutation | 3 | |
| ERBB2 | SKCM | chr17 | 37871608 | 37871608 | C | T | Silent | p.I376I | 3 |
| ERBB2 | UCEC | chr17 | 37864650 | 37864650 | T | G | Missense_Mutation | p.I101S | 3 |
| ERBB2 | BLCA | chr17 | 37879583 | 37879583 | C | G | Missense_Mutation | 3 | |
| ERBB2 | BLCA | chr17 | 37866444 | 37866444 | C | T | Missense_Mutation | 3 | |
| ERBB2 | STAD | chr17 | 37883212 | 37883212 | G | A | Missense_Mutation | p.A1039T | 3 |
| ERBB2 | BLCA | chr17 | 37879588 | 37879588 | A | G | Missense_Mutation | 3 | |
| ERBB2 | KIRP | chr17 | 37876080 | 37876080 | A | C | Silent | p.R647R | 3 |
| ERBB2 | BRCA | chr17 | 37868205 | 37868205 | G | C | Missense_Mutation | p.G309A | 3 |
| ERBB2 | CHOL | chr17 | 37880219 | 37880219 | T | C | Silent | 3 | |
| ERBB2 | BLCA | chr17 | 37864656 | 37864656 | G | A | Missense_Mutation | p.R73Q | 2 |
| ERBB2 | HNSC | chr17 | 37865676 | 37865676 | C | T | Missense_Mutation | p.T152I | 2 |
| ERBB2 | ESCA | chr17 | 37881021 | 37881021 | C | T | Missense_Mutation | 2 | |
| ERBB2 | SKCM | chr17 | 37883082 | 37883082 | C | T | Silent | p.G965G | 2 |
| ERBB2 | PRAD | chr17 | 37881014 | 37881014 | T | C | Silent | p.Y751Y | 2 |
| ERBB2 | ESCA | chr17 | 37879610 | 37879610 | T | A | Missense_Mutation | p.L662Q | 2 |
| ERBB2 | ESCA | chr17 | 37881323 | 37881323 | G | A | Missense_Mutation | 2 | |
| ERBB2 | BLCA | chr17 | 37881392 | 37881392 | A | G | Missense_Mutation | p.T862A | 2 |
| ERBB2 | THYM | chr17 | 37881427 | 37881427 | C | T | Silent | 2 | |
| ERBB2 | LUAD | chr17 | 37880981 | 37880982 | - | GCATACGTGATG | In_Frame_Ins | p.774_775insAYVM | 2 |
| ERBB2 | CHOL | chr17 | 37880219 | 37880219 | T | C | Silent | p.L725L | 2 |
| ERBB2 | PAAD | chr17 | 37864607 | 37864607 | G | A | Missense_Mutation | 2 | |
| ERBB2 | SKCM | chr17 | 37882045 | 37882045 | G | A | Silent | p.K907K | 2 |
| ERBB2 | SKCM | chr17 | 37882009 | 37882009 | G | A | Silent | p.G895G | 2 |
| ERBB2 | BLCA | chr17 | 37879658 | 37879658 | G | A | Missense_Mutation | 2 | |
| ERBB2 | ESCA | chr17 | 37881323 | 37881323 | G | A | Missense_Mutation | p.V839M | 2 |
| ERBB2 | BLCA | chr17 | 37866359 | 37866359 | G | T | Missense_Mutation | p.G222C | 2 |
| ERBB2 | HNSC | chr17 | 37881982 | 37881982 | G | A | Missense_Mutation | p.M916I | 2 |
| ERBB2 | PAAD | chr17 | 37868294 | 37868294 | G | A | Missense_Mutation | 2 | |
| ERBB2 | SKCM | chr17 | 37864763 | 37864763 | G | A | Missense_Mutation | p.E109K | 2 |
| ERBB2 | SKCM | chr17 | 37879848 | 37879848 | C | T | Silent | p.L685L | 2 |
| ERBB2 | CESC | chr17 | 37866407 | 37866407 | G | A | Missense_Mutation | 2 | |
| ERBB2 | THYM | chr17 | 37881075 | 37881075 | C | T | Missense_Mutation | 2 | |
| ERBB2 | BLCA | chr17 | 37880257 | 37880257 | C | G | Missense_Mutation | 2 | |
| ERBB2 | LGG | chr17 | 37866098 | 37866098 | C | T | Missense_Mutation | p.R173C | 2 |
| ERBB2 | LUAD | chr17 | 37884217 | 37884218 | - | G | Frame_Shift_Ins | p.R1230fs | 2 |
| ERBB2 | ESCA | chr17 | 37865579 | 37865579 | A | G | Missense_Mutation | p.K150E | 2 |
| ERBB2 | UCEC | chr17 | 37866082 | 37866082 | G | A | Silent | p.P197 | 2 |
| ERBB2 | BRCA | chr17 | 37872089 | 37872089 | T | G | Missense_Mutation | p.H470Q | 2 |
| ERBB2 | HNSC | chr17 | 37884229 | 37884229 | C | T | Missense_Mutation | p.P1204S | 2 |
| ERBB2 | BLCA | chr17 | 37881392 | 37881392 | A | G | Missense_Mutation | p.T832A | 2 |
| ERBB2 | SKCM | chr17 | 37872083 | 37872083 | C | T | Silent | p.I438I | 2 |
| ERBB2 | CESC | chr17 | 37884255 | 37884255 | G | A | Silent | 2 | |
| ERBB2 | BLCA | chr17 | 37866671 | 37866671 | G | C | Missense_Mutation | 2 | |
| ERBB2 | UCEC | chr17 | 37879820 | 37879820 | G | A | Silent | p.A705 | 2 |
| ERBB2 | BRCA | chr17 | 37880257 | 37880257 | C | G | Missense_Mutation | p.I767M | 2 |
| ERBB2 | SKCM | chr17 | 37866434 | 37866434 | C | T | Missense_Mutation | p.P217S | 2 |
| ERBB2 | PAAD | chr17 | 37883966 | 37883966 | G | A | Missense_Mutation | 2 | |
| ERBB2 | SKCM | chr17 | 37864606 | 37864606 | C | T | Silent | p.I56I | 2 |
| ERBB2 | THYM | chr17 | 37881075 | 37881075 | C | T | Missense_Mutation | p.P802S | 2 |
| ERBB2 | ESCA | chr17 | 37881021 | 37881021 | C | T | Missense_Mutation | p.R784C | 2 |
| ERBB2 | STAD | chr17 | 37880998 | 37880998 | G | T | Missense_Mutation | p.G776V | 2 |
| ERBB2 | UCEC | chr17 | 37880207 | 37880207 | G | A | Missense_Mutation | p.A751T | 2 |
| ERBB2 | BLCA | chr17 | 37864656 | 37864656 | G | A | Missense_Mutation | 2 | |
| ERBB2 | SKCM | chr17 | 37864760 | 37864760 | C | T | Missense_Mutation | p.R108W | 2 |
| ERBB2 | TGCT | chr17 | 37879626 | 37879626 | G | C | Missense_Mutation | 2 | |
| ERBB2 | PAAD | chr17 | 37864607 | 37864607 | G | A | Missense_Mutation | p.A87T | 2 |
| ERBB2 | SKCM | chr17 | 37884092 | 37884092 | G | A | Missense_Mutation | p.G1158E | 2 |
| ERBB2 | THYM | chr17 | 37865600 | 37865600 | C | T | Missense_Mutation | p.R157W | 2 |
| ERBB2 | LGG | chr17 | 37866098 | 37866098 | C | T | Missense_Mutation | 2 | |
| ERBB2 | ESCA | chr17 | 37866417 | 37866417 | C | T | Missense_Mutation | 2 | |
| ERBB2 | STAD | chr17 | 37863285 | 37863285 | C | T | Missense_Mutation | p.P39L | 2 |
| ERBB2 | UCEC | chr17 | 37881400 | 37881400 | C | T | Silent | p.F864 | 2 |
| ERBB2 | BLCA | chr17 | 37866662 | 37866662 | G | T | Missense_Mutation | p.D247Y | 2 |
| ERBB2 | BRCA | chr17 | 37883561 | 37883561 | A | C | Missense_Mutation | p.D1058A | 2 |
| ERBB2 | PAAD | chr17 | 37882024 | 37882024 | G | T | Missense_Mutation | p.E930D | 2 |
| ERBB2 | SKCM | chr17 | 37866711 | 37866711 | C | T | Missense_Mutation | p.A263V | 2 |
| ERBB2 | BLCA | chr17 | 37880257 | 37880257 | C | G | Missense_Mutation | p.I737M | 2 |
| ERBB2 | CESC | chr17 | 37879603 | 37879603 | G | C | Missense_Mutation | 2 | |
| ERBB2 | ESCA | chr17 | 37879610 | 37879610 | T | A | Missense_Mutation | 2 | |
| ERBB2 | STAD | chr17 | 37864636 | 37864636 | G | A | Silent | p.L96L | 2 |
| ERBB2 | UCEC | chr17 | 37881620 | 37881620 | G | A | Missense_Mutation | p.R897Q | 2 |
| ERBB2 | PAAD | chr17 | 37883966 | 37883966 | G | A | Missense_Mutation | p.R1146Q | 2 |
| ERBB2 | SKCM | chr17 | 37881310 | 37881310 | C | T | Silent | p.S804S | 2 |
| ERBB2 | CESC | chr17 | 37881425 | 37881425 | G | A | Missense_Mutation | 2 | |
| ERBB2 | DLBC | chr17 | 37879588 | 37879588 | A | G | Missense_Mutation | p.I655V | 2 |
| ERBB2 | STAD | chr17 | 37879601 | 37879601 | T | A | Missense_Mutation | 2 | |
| ERBB2 | KIRP | chr17 | 37883597 | 37883597 | C | T | Missense_Mutation | p.A1070V | 2 |
| ERBB2 | HNSC | chr17 | 37883113 | 37883113 | C | T | Missense_Mutation | p.R976C | 2 |
| ERBB2 | ESCA | chr17 | 37865579 | 37865579 | A | G | Missense_Mutation | 2 | |
| ERBB2 | STAD | chr17 | 37873670 | 37873670 | C | A | Missense_Mutation | p.P612H | 2 |
| ERBB2 | UCEC | chr17 | 37884261 | 37884261 | G | T | Missense_Mutation | p.E1244D | 2 |
| ERBB2 | BLCA | chr17 | 37883949 | 37883949 | G | A | Silent | p.V1110V | 2 |
| ERBB2 | LIHC | chr17 | 37883733 | 37883733 | C | - | Frame_Shift_Del | p.D1085fs | 2 |
| ERBB2 | SKCM | chr17 | 37883074 | 37883074 | G | A | Missense_Mutation | p.D963N | 2 |
| ERBB2 | CESC | chr17 | 37871593 | 37871593 | G | C | Missense_Mutation | 2 | |
| ERBB2 | ESCA | chr17 | 37866417 | 37866417 | C | T | Missense_Mutation | p.A241V | 2 |
| ERBB2 | STAD | chr17 | 37883212 | 37883212 | G | A | Missense_Mutation | 2 | |
| ERBB2 | BLCA | chr17 | 37866352 | 37866352 | C | A | Silent | 1 | |
| ERBB2 | LIHC | chr17 | 37884188 | 37884188 | T | C | Missense_Mutation | 1 | |
| ERBB2 | OV | chr17 | 35121734 | 35121734 | C | T | Missense_Mutation | 1 | |
| ERBB2 | SARC | chr17 | 37872823 | 37872823 | C | T | Nonsense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37879820 | 37879820 | G | A | Silent | p.A705A | 1 |
| ERBB2 | CHOL | chr17 | 37883077 | 37883077 | T | G | Missense_Mutation | 1 | |
| ERBB2 | STAD | chr17 | 37883212 | 37883212 | G | A | Missense_Mutation | p.A1009T | 1 |
| ERBB2 | HNSC | chr17 | 37872674 | 37872674 | G | A | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37880261 | 37880261 | G | C | Missense_Mutation | p.D739H | 1 |
| ERBB2 | COAD | chr17 | 37879595 | 37879595 | C | T | Missense_Mutation | p.A627V | 1 |
| ERBB2 | SKCM | chr17 | 37883679 | 37883679 | G | A | Silent | p.G1067G | 1 |
| ERBB2 | ESCA | chr17 | 37880988 | 37880989 | - | TGA | In_Frame_Ins | p.742_743ins* | 1 |
| ERBB2 | TGCT | chr17 | 37879626 | 37879626 | G | C | Missense_Mutation | p.L637F | 1 |
| ERBB2 | BLCA | chr17 | 37873620 | 37873620 | C | A | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37880997 | 37880997 | G | A | Missense_Mutation | p.G746S | 1 |
| ERBB2 | KICH | chr17 | 37881117 | 37881117 | C | T | Missense_Mutation | 1 | |
| ERBB2 | LIHC | chr17 | 37868594 | 37868594 | G | - | Frame_Shift_Del | p.M317fs | 1 |
| ERBB2 | PAAD | chr17 | 37864607 | 37864607 | G | A | Missense_Mutation | p.A57T | 1 |
| ERBB2 | LUAD | chr17 | 37865605 | 37865605 | C | T | Silent | p.N158N | 1 |
| ERBB2 | BLCA | chr17 | 37871731 | 37871731 | C | T | Silent | p.L389L | 1 |
| ERBB2 | CESC | chr17 | 37883623 | 37883623 | G | G | Missense_Mutation | 1 | |
| ERBB2 | STAD | chr17 | 37872036 | 37872036 | C | A | Missense_Mutation | 1 | |
| ERBB2 | HNSC | chr17 | 37868215 | 37868215 | C | G | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37884048 | 37884048 | C | G | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37872839 | 37872839 | C | T | Missense_Mutation | p.S543L | 1 |
| ERBB2 | KIRP | chr17 | 37883597 | 37883597 | C | T | Missense_Mutation | p.A1040V | 1 |
| ERBB2 | BLCA | chr17 | 37871714 | 37871714 | C | T | Missense_Mutation | 1 | |
| ERBB2 | HNSC | chr17 | 37879629 | 37879629 | G | T | Silent | p.G638G | 1 |
| ERBB2 | LIHC | chr17 | 37872839 | 37872839 | C | A | Nonsense_Mutation | p.S573X | 1 |
| ERBB2 | OV | chr17 | 35134901 | 35134901 | C | G | Missense_Mutation | p.P856R | 1 |
| ERBB2 | SARC | chr17 | 37866093 | 37866093 | G | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37864656 | 37864656 | G | A | Missense_Mutation | p.R103Q | 1 |
| ERBB2 | ESCA | chr17 | 37865644 | 37865644 | C | T | Silent | 1 | |
| ERBB2 | STAD | chr17 | 37879601 | 37879601 | T | A | Missense_Mutation | p.V629D | 1 |
| ERBB2 | PAAD | chr17 | 37882024 | 37882024 | G | T | Missense_Mutation | p.E900D | 1 |
| ERBB2 | SKCM | chr17 | 37865597 | 37865597 | C | T | Nonsense_Mutation | p.Q126X | 1 |
| ERBB2 | BLCA | chr17 | 37865656 | 37865656 | G | A | Silent | p.K145K | 1 |
| ERBB2 | COAD | chr17 | 37879688 | 37879688 | G | A | Missense_Mutation | p.R658Q | 1 |
| ERBB2 | SKCM | chr17 | 37883678 | 37883678 | G | A | Missense_Mutation | p.G1067E | 1 |
| ERBB2 | GBM | chr17 | 37884124 | 37884124 | C | A | Missense_Mutation | p.P1199T | 1 |
| ERBB2 | THCA | chr17 | 37882044 | 37882044 | A | G | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37881979 | 37881979 | G | C | Silent | 1 | |
| ERBB2 | KIRC | chr17 | 37866350 | 37866350 | G | A | Missense_Mutation | p.V219I | 1 |
| ERBB2 | LIHC | chr17 | 37881654 | 37881654 | T | - | Splice_Site | p.Y878_splice | 1 |
| ERBB2 | KIRP | chr17 | 37883164 | 37883164 | T | - | Frame_Shift_Del | p.Y1023fs | 1 |
| ERBB2 | LUAD | chr17 | 37865605 | 37865605 | C | T | Silent | p.N128N | 1 |
| ERBB2 | LUAD | chr17 | 37871593 | 37871593 | G | A | Silent | p.E401E | 1 |
| ERBB2 | PRAD | chr17 | 37872561 | 37872561 | G | - | Frame_Shift_Del | p.E477fs | 1 |
| ERBB2 | BLCA | chr17 | 37884259 | 37884259 | G | C | Missense_Mutation | p.E1244Q | 1 |
| ERBB2 | CESC | chr17 | 37883117 | 37883117 | C | C | Missense_Mutation | 1 | |
| ERBB2 | ESCA | chr17 | 37880988 | 37880989 | - | TGA | In_Frame_Ins | p.775in_frame_insM | 1 |
| ERBB2 | STAD | chr17 | 37872036 | 37872036 | C | A | Missense_Mutation | p.L453M | 1 |
| ERBB2 | HNSC | chr17 | 37882022 | 37882022 | G | A | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37884180 | 37884180 | C | T | Silent | 1 | |
| ERBB2 | STAD | chr17 | 37879583 | 37879583 | C | T | Missense_Mutation | p.S623F | 1 |
| ERBB2 | HNSC | chr17 | 37882022 | 37882022 | G | A | Missense_Mutation | p.E900K | 1 |
| ERBB2 | BLCA | chr17 | 37866352 | 37866352 | C | A | Silent | p.V189V | 1 |
| ERBB2 | HNSC | chr17 | 37868215 | 37868215 | C | G | Silent | p.T282T | 1 |
| ERBB2 | LIHC | chr17 | 37876052 | 37876052 | G | A | Silent | p.L637L | 1 |
| ERBB2 | OV | chr17 | 35121734 | 35121734 | C | T | Missense_Mutation | p.S310F | 1 |
| ERBB2 | SARC | chr17 | 37864783 | 37864783 | C | T | Silent | p.L145 | 1 |
| ERBB2 | CHOL | chr17 | 37883077 | 37883077 | T | G | Missense_Mutation | p.L994V | 1 |
| ERBB2 | PAAD | chr17 | 37883966 | 37883966 | G | A | Missense_Mutation | p.R1116Q | 1 |
| ERBB2 | BLCA | chr17 | 37884151 | 37884151 | C | T | Missense_Mutation | p.H1178Y | 1 |
| ERBB2 | COAD | chr17 | 37883088 | 37883088 | C | A | Silent | p.A967A | 1 |
| ERBB2 | SKCM | chr17 | 37866370 | 37866370 | C | T | Silent | p.A195A | 1 |
| ERBB2 | GBM | chr17 | 37866667 | 37866667 | G | A | Silent | p.T278T | 1 |
| ERBB2 | BLCA | chr17 | 37883134 | 37883134 | G | C | Missense_Mutation | 1 | |
| ERBB2 | KIRC | chr17 | 37871549 | 37871550 | - | C | Frame_Shift_Ins | p.S387fs | 1 |
| ERBB2 | LIHC | chr17 | 37883774 | 37883774 | C | - | Frame_Shift_Del | p.A1099fs | 1 |
| ERBB2 | KIRP | chr17 | 37876080 | 37876080 | A | C | Silent | 1 | |
| ERBB2 | LUAD | chr17 | 37871593 | 37871593 | G | A | Silent | p.E371E | 1 |
| ERBB2 | READ | chr17 | 37879794 | 37879794 | G | T | Missense_Mutation | p.V667L | 1 |
| ERBB2 | BLCA | chr17 | 37872839 | 37872839 | C | T | Missense_Mutation | p.S573L | 1 |
| ERBB2 | CESC | chr17 | 37866407 | 37866407 | G | A | Missense_Mutation | p.E238K | 1 |
| ERBB2 | STAD | chr17 | 37879583 | 37879583 | C | T | Missense_Mutation | p.S653F | 1 |
| ERBB2 | HNSC | chr17 | 37883561 | 37883561 | A | C | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37879903 | 37879903 | C | T | Missense_Mutation | 1 | |
| ERBB2 | ESCA | chr17 | 37880988 | 37880989 | - | TGA | In_Frame_Ins | p.V773delinsVM | 1 |
| ERBB2 | STAD | chr17 | 37872036 | 37872036 | C | A | Missense_Mutation | p.L423M | 1 |
| ERBB2 | HNSC | chr17 | 37866346 | 37866346 | C | T | Silent | p.R187R | 1 |
| ERBB2 | BLCA | chr17 | 37866078 | 37866078 | C | T | Missense_Mutation | p.S166F | 1 |
| ERBB2 | HNSC | chr17 | 37880180 | 37880180 | G | C | Missense_Mutation | p.D742H | 1 |
| ERBB2 | LIHC | chr17 | 37882886 | 37882886 | G | T | Missense_Mutation | p.D982Y | 1 |
| ERBB2 | BLCA | chr17 | 37868200 | 37868200 | C | T | Silent | p.D307D | 1 |
| ERBB2 | PAAD | chr17 | 37863277 | 37863277 | T | A | Silent | p.P36P | 1 |
| ERBB2 | BLCA | chr17 | 37868682 | 37868682 | C | T | Silent | p.L347L | 1 |
| ERBB2 | BRCA | chr17 | 37879581 | 37879581 | G | T | Silent | p.T652 | 1 |
| ERBB2 | COAD | chr17 | 37883769 | 37883769 | C | T | Silent | p.Y1097Y | 1 |
| ERBB2 | SKCM | chr17 | 37884091 | 37884091 | G | A | Missense_Mutation | p.G1158R | 1 |
| ERBB2 | GBM | chr17 | 37884093 | 37884093 | G | T | Silent | 1 | |
| ERBB2 | THYM | chr17 | 37863262 | 37863262 | G | T | Translation_Start_Site | 1 | |
| ERBB2 | BLCA | chr17 | 37864603 | 37864603 | C | G | Silent | 1 | |
| ERBB2 | KIRC | chr17 | 37876087 | 37876087 | G | C | Missense_Mutation | p.S649T | 1 |
| ERBB2 | LIHC | chr17 | 37863340 | 37863340 | G | - | Frame_Shift_Del | p.Q27fs | 1 |
| ERBB2 | KIRP | chr17 | 37883597 | 37883597 | C | T | Missense_Mutation | 1 | |
| ERBB2 | LUAD | chr17 | 37866373 | 37866373 | C | T | Silent | p.R196R | 1 |
| ERBB2 | LUAD | chr17 | 37880997 | 37880998 | - | TCT | In_Frame_Ins | p.776_776G>VC | 1 |
| ERBB2 | READ | chr17 | 37880998 | 37880998 | G | T | Missense_Mutation | p.G746V | 1 |
| ERBB2 | CESC | chr17 | 37866671 | 37866671 | G | C | Missense_Mutation | p.E280Q | 1 |
| ERBB2 | STAD | chr17 | 37868208 | 37868208 | C | A | Missense_Mutation | p.S310Y | 1 |
| ERBB2 | HNSC | chr17 | 37879629 | 37879629 | G | T | Silent | 1 | |
| ERBB2 | UCEC | chr17 | 37884218 | 37884218 | G | - | Frame_Shift_Del | p.R1230fs | 1 |
| ERBB2 | BLCA | chr17 | 37879820 | 37879820 | G | A | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37884180 | 37884180 | C | T | Silent | p.F1187F | 1 |
| ERBB2 | CHOL | chr17 | 37883077 | 37883077 | T | G | Missense_Mutation | p.L964V | 1 |
| ERBB2 | ESCA | chr17 | 37866417 | 37866417 | C | T | Missense_Mutation | p.A211V | 1 |
| ERBB2 | STAD | chr17 | 37856540 | 37856540 | C | - | Frame_Shift_Del | p.P18fs | 1 |
| ERBB2 | HNSC | chr17 | 37883982 | 37883982 | G | A | Silent | p.S1121S | 1 |
| ERBB2 | BLCA | chr17 | 37866359 | 37866359 | G | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37871714 | 37871714 | C | T | Missense_Mutation | p.S383L | 1 |
| ERBB2 | LIHC | chr17 | 37868282 | 37868282 | A | G | Missense_Mutation | p.S335G | 1 |
| ERBB2 | PAAD | chr17 | 37868294 | 37868294 | G | A | Missense_Mutation | p.A339T | 1 |
| ERBB2 | BLCA | chr17 | 37873620 | 37873620 | C | A | Missense_Mutation | p.F565L | 1 |
| ERBB2 | COAD | chr17 | 37884208 | 37884208 | C | T | Missense_Mutation | p.P1197S | 1 |
| ERBB2 | SKCM | chr17 | 37873615 | 37873615 | C | T | Missense_Mutation | p.P564S | 1 |
| ERBB2 | GBM | chr17 | 37866667 | 37866667 | G | A | Silent | 1 | |
| ERBB2 | KIRC | chr17 | 37876087 | 37876087 | G | C | Splice_Site | p.S619_splice | 1 |
| ERBB2 | LIHC | chr17 | 37864686 | 37864686 | C | - | Frame_Shift_Del | p.A83fs | 1 |
| ERBB2 | LUAD | chr17 | 37866340 | 37866340 | G | T | Splice_Site | p.L185_splice | 1 |
| ERBB2 | READ | chr17 | 37882854 | 37882854 | A | G | Missense_Mutation | p.E941G | 1 |
| ERBB2 | CESC | chr17 | 37879603 | 37879603 | G | C | Missense_Mutation | p.G660R | 1 |
| ERBB2 | HNSC | chr17 | 37865676 | 37865676 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37872107 | 37872107 | C | T | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37884048 | 37884048 | C | G | Silent | p.L1143L | 1 |
| ERBB2 | COAD | chr17 | 37864666 | 37864666 | G | A | Silent | p.Q76Q | 1 |
| ERBB2 | ESCA | chr17 | 37879610 | 37879610 | T | A | Missense_Mutation | p.L632Q | 1 |
| ERBB2 | STAD | chr17 | 37882863 | 37882863 | C | T | Missense_Mutation | p.S974F | 1 |
| ERBB2 | HNSC | chr17 | 37882022 | 37882022 | G | A | Missense_Mutation | p.E930K | 1 |
| ERBB2 | LIHC | chr17 | 37872630 | 37872630 | C | T | Silent | p.N530N | 1 |
| ERBB2 | PAAD | chr17 | 37882024 | 37882024 | G | T | Missense_Mutation | 1 | |
| ERBB2 | PCPG | chr17 | 37880178 | 37880178 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37881979 | 37881979 | G | C | Silent | p.L885L | 1 |
| ERBB2 | COAD | chr17 | 37881621 | 37881621 | G | A | Silent | p.R867R | 1 |
| ERBB2 | SKCM | chr17 | 37871598 | 37871598 | T | C | Missense_Mutation | p.L373P | 1 |
| ERBB2 | GBM | chr17 | 37872844 | 37872844 | A | T | Missense_Mutation | 1 | |
| ERBB2 | THYM | chr17 | 37865600 | 37865600 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37884259 | 37884259 | G | C | Missense_Mutation | 1 | |
| ERBB2 | KIRC | chr17 | 37882078 | 37882078 | C | A | Silent | p.T918T | 1 |
| ERBB2 | LIHC | chr17 | 37865605 | 37865605 | C | - | Frame_Shift_Del | p.N128fs | 1 |
| ERBB2 | LGG | chr17 | 37876049 | 37876049 | C | T | Silent | p.D606D | 1 |
| ERBB2 | LUAD | chr17 | 37880997 | 37880998 | - | TAT | In_Frame_Ins | p.745_746insY | 1 |
| ERBB2 | LUSC | chr17 | 37883158 | 37883158 | G | C | Missense_Mutation | p.E1021Q | 1 |
| ERBB2 | SARC | chr17 | 37864783 | 37864783 | C | T | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37868216 | 37868216 | C | G | Missense_Mutation | p.L313V | 1 |
| ERBB2 | CESC | chr17 | 37881425 | 37881425 | G | A | Missense_Mutation | p.D873N | 1 |
| ERBB2 | ESCA | chr17 | 37866342 | 37866342 | C | T | Missense_Mutation | p.T216M | 1 |
| ERBB2 | STAD | chr17 | 37879601 | 37879601 | T | A | Missense_Mutation | p.V659D | 1 |
| ERBB2 | HNSC | chr17 | 37884229 | 37884229 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37863257 | 37863257 | G | C | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37879903 | 37879903 | C | T | Missense_Mutation | p.T703I | 1 |
| ERBB2 | ESCA | chr17 | 37881000 | 37881000 | G | A | Missense_Mutation | p.V747M | 1 |
| ERBB2 | STAD | chr17 | 37866592 | 37866592 | G | T | Splice_Site | 1 | |
| ERBB2 | HNSC | chr17 | 37880180 | 37880180 | G | C | Missense_Mutation | p.D712H | 1 |
| ERBB2 | BLCA | chr17 | 37880997 | 37880997 | G | A | Missense_Mutation | 1 | |
| ERBB2 | HNSC | chr17 | 37883578 | 37883578 | G | C | Missense_Mutation | p.E1064Q | 1 |
| ERBB2 | LIHC | chr17 | 37863315 | 37863315 | T | C | Missense_Mutation | p.L49P | 1 |
| ERBB2 | PRAD | chr17 | 37881014 | 37881014 | T | C | Silent | p.Y781Y | 1 |
| ERBB2 | BLCA | chr17 | 37883134 | 37883134 | G | C | Missense_Mutation | p.D983H | 1 |
| ERBB2 | CESC | chr17 | 37882863 | 37882863 | C | T | Missense_Mutation | 1 | |
| ERBB2 | COAD | chr17 | 37883787 | 37883787 | C | T | Silent | p.C1103C | 1 |
| ERBB2 | SKCM | chr17 | 37880219 | 37880219 | T | A | Missense_Mutation | p.L725M | 1 |
| ERBB2 | GBM | chr17 | 37879617 | 37879617 | C | T | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37872839 | 37872839 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37881132 | 37881132 | G | C | Missense_Mutation | 1 | |
| ERBB2 | KIRC | chr17 | 37866350 | 37866350 | G | A | Missense_Mutation | p.V189I | 1 |
| ERBB2 | LIHC | chr17 | 37881985 | 37881985 | T | - | Frame_Shift_Del | p.T887fs | 1 |
| ERBB2 | LGG | chr17 | 37865570 | 37865570 | G | T | Splice_Site | 1 | |
| ERBB2 | LUSC | chr17 | 37864713 | 37864713 | C | T | Missense_Mutation | p.P122L | 1 |
| ERBB2 | SARC | chr17 | 37883121 | 37883121 | G | T | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37880261 | 37880261 | G | A | Missense_Mutation | p.D769N | 1 |
| ERBB2 | CESC | chr17 | 37871593 | 37871593 | G | C | Missense_Mutation | p.E401D | 1 |
| ERBB2 | HNSC | chr17 | 37883113 | 37883113 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37879820 | 37879820 | G | A | Silent | p.A675A | 1 |
| ERBB2 | ESCA | chr17 | 37865579 | 37865579 | A | G | Missense_Mutation | p.K120E | 1 |
| ERBB2 | HNSC | chr17 | 37872674 | 37872674 | G | A | Missense_Mutation | p.R515Q | 1 |
| ERBB2 | BLCA | chr17 | 37866662 | 37866662 | G | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37865593 | 37865593 | G | C | Missense_Mutation | p.L124F | 1 |
| ERBB2 | HNSC | chr17 | 37879629 | 37879629 | G | T | Silent | p.G668G | 1 |
| ERBB2 | PRAD | chr17 | 37883719 | 37883719 | C | T | Missense_Mutation | p.R1081W | 1 |
| ERBB2 | BLCA | chr17 | 37864603 | 37864603 | C | G | Silent | p.L55L | 1 |
| ERBB2 | CESC | chr17 | 37865659 | 37865659 | C | T | Silent | 1 | |
| ERBB2 | DLBC | chr17 | 37872035 | 37872035 | G | T | Missense_Mutation | p.W452C | 1 |
| ERBB2 | SKCM | chr17 | 37866686 | 37866686 | C | T | Missense_Mutation | p.P255S | 1 |
| ERBB2 | GBM | chr17 | 37879682 | 37879682 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37871731 | 37871731 | C | T | Silent | 1 | |
| ERBB2 | KIRC | chr17 | 37871549 | 37871550 | - | C | Frame_Shift_Ins | p.Q357fs | 1 |
| ERBB2 | LIHC | chr17 | 37881989 | 37881989 | G | - | Frame_Shift_Del | p.G889fs | 1 |
| ERBB2 | LUSC | chr17 | 37866733 | 37866733 | C | T | Silent | p.Y301_splice | 1 |
| ERBB2 | SARC | chr17 | 37884037 | 37884037 | C | G | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37884048 | 37884048 | C | G | Silent | p.L1173L | 1 |
| ERBB2 | CESC | chr17 | 37883623 | 37883623 | G | A | Missense_Mutation | p.E1079K | 1 |
| ERBB2 | HNSC | chr17 | 37866346 | 37866346 | C | T | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37872107 | 37872107 | C | T | Silent | p.F446F | 1 |
| ERBB2 | SKCM | chr17 | 37863355 | 37863355 | C | T | Silent | p.L32L | 1 |
| ERBB2 | ESCA | chr17 | 37881021 | 37881021 | C | T | Missense_Mutation | p.R754C | 1 |
| ERBB2 | TGCT | chr17 | 37879626 | 37879626 | G | C | Missense_Mutation | p.L667F | 1 |
| ERBB2 | BLCA | chr17 | 37881041 | 37881041 | G | A | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37866359 | 37866359 | G | T | Missense_Mutation | p.G192C | 1 |
| ERBB2 | HNSC | chr17 | 37871770 | 37871770 | C | T | Missense_Mutation | p.R432W | 1 |
| ERBB2 | LIHC | chr17 | 37882058 | 37882058 | C | - | Frame_Shift_Del | p.P912fs | 1 |
| ERBB2 | LUAD | chr17 | 37880997 | 37880998 | - | TCT | In_Frame_Ins | p.745_746insS | 1 |
| ERBB2 | PRAD | chr17 | 37881050 | 37881050 | G | A | Silent | p.T763T | 1 |
| ERBB2 | DLBC | chr17 | 37863268 | 37863268 | G | A | Silent | p.L33L | 1 |
| ERBB2 | SKCM | chr17 | 37865597 | 37865597 | C | T | Nonsense_Mutation | p.Q126* | 1 |
| ERBB2 | GBM | chr17 | 37881372 | 37881372 | G | T | Missense_Mutation | 1 | |
| ERBB2 | THYM | chr17 | 37881427 | 37881427 | C | T | Silent | p.D873D | 1 |
| ERBB2 | LUAD | chr17 | 37876038 | 37876038 | A | T | Splice_Site | 1 | |
| ERBB2 | BLCA | chr17 | 37872618 | 37872618 | C | G | Silent | p.T496T | 1 |
| ERBB2 | HNSC | chr17 | 37882904 | 37882904 | G | A | Missense_Mutation | p.V958I | 1 |
| ERBB2 | LGG | chr17 | 37880224 | 37880224 | G | T | Missense_Mutation | 1 | |
| ERBB2 | OV | chr17 | 35134658 | 35134658 | G | T | Missense_Mutation | 1 | |
| ERBB2 | SARC | chr17 | 37864783 | 37864783 | C | T | Silent | p.L115L | 1 |
| ERBB2 | BLCA | chr17 | 37884180 | 37884180 | C | T | Silent | p.F1217F | 1 |
| ERBB2 | CESC | chr17 | 37884255 | 37884255 | G | A | Silent | p.T1242 | 1 |
| ERBB2 | HNSC | chr17 | 37883982 | 37883982 | G | A | Silent | 1 | |
| ERBB2 | BLCA | chr17 | 37881041 | 37881041 | G | A | Silent | p.L760L | 1 |
| ERBB2 | SKCM | chr17 | 37863388 | 37863388 | C | T | Silent | p.F43F | 1 |
| ERBB2 | ESCA | chr17 | 37866342 | 37866342 | C | T | Missense_Mutation | p.T186M | 1 |
| ERBB2 | TGCT | chr17 | 37884152 | 37884152 | A | C | Missense_Mutation | p.H1178P | 1 |
| ERBB2 | BLCA | chr17 | 37865656 | 37865656 | G | A | Silent | 1 | |
| ERBB2 | HNSC | chr17 | 37865676 | 37865676 | C | T | Missense_Mutation | p.T182I | 1 |
| ERBB2 | LIHC | chr17 | 37882064 | 37882064 | C | - | Frame_Shift_Del | p.P915fs | 1 |
| ERBB2 | LUAD | chr17 | 37884217 | 37884218 | - | G | Frame_Shift_Ins | p.G1200fs | 1 |
| ERBB2 | PRAD | chr17 | 37884170 | 37884170 | G | A | Missense_Mutation | p.S1184N | 1 |
| ERBB2 | BLCA | chr17 | 37866671 | 37866671 | G | C | Missense_Mutation | p.E250Q | 1 |
| ERBB2 | GBM | chr17 | 37884124 | 37884124 | C | A | Missense_Mutation | 1 | |
| ERBB2 | THYM | chr17 | 37881075 | 37881075 | C | T | Missense_Mutation | p.P772S | 1 |
| ERBB2 | BLCA | chr17 | 37868216 | 37868216 | C | G | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37866444 | 37866444 | C | T | Missense_Mutation | p.S220F | 1 |
| ERBB2 | LUAD | chr17 | 37879576 | 37879576 | C | G | Missense_Mutation | p.L621V | 1 |
| ERBB2 | BLCA | chr17 | 37866078 | 37866078 | C | T | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37863257 | 37863257 | G | C | Missense_Mutation | p.D30H | 1 |
| ERBB2 | LIHC | chr17 | 37872164 | 37872164 | C | T | Silent | 1 | |
| ERBB2 | OV | chr17 | 35135143 | 35135143 | G | A | Missense_Mutation | 1 | |
| ERBB2 | SARC | chr17 | 37876039 | 37876039 | G | T | Splice_Site | 1 | |
| ERBB2 | BLCA | chr17 | 37879903 | 37879903 | C | T | Missense_Mutation | p.T733I | 1 |
| ERBB2 | HNSC | chr17 | 37882904 | 37882904 | G | A | Missense_Mutation | 1 | |
| ERBB2 | BLCA | chr17 | 37880261 | 37880261 | G | T | Missense_Mutation | p.D739Y | 1 |
| ERBB2 | COAD | chr17 | 37864776 | 37864776 | G | A | Missense_Mutation | p.R113Q | 1 |
| ERBB2 | SKCM | chr17 | 37876074 | 37876074 | G | A | Missense_Mutation | p.E615K | 1 |
| ERBB2 | ESCA | chr17 | 37881323 | 37881323 | G | A | Missense_Mutation | p.V809M | 1 |
| ERBB2 | TGCT | chr17 | 37879599 | 37879599 | G | T | Silent | p.V628V | 1 |
| ERBB2 | BLCA | chr17 | 37884151 | 37884151 | C | T | Missense_Mutation | 1 | |
| ERBB2 | HNSC | chr17 | 37884229 | 37884229 | C | T | Missense_Mutation | p.P1234S | 1 |
| ERBB2 | PAAD | chr17 | 37868294 | 37868294 | G | A | Missense_Mutation | p.A309T | 1 |
| ERBB2 | LUAD | chr17 | 37876038 | 37876038 | A | T | Splice_Site | p.S633_splice | 1 |
| ERBB2 | PRAD | chr17 | 37866372 | 37866372 | G | A | Missense_Mutation | p.R196H | 1 |
| ERBB2 | BLCA | chr17 | 37881132 | 37881132 | G | C | Missense_Mutation | p.D791H | 1 |
| ERBB2 | HNSC | chr17 | 37880180 | 37880180 | G | C | Missense_Mutation | 1 | |
| ERBB2 | THYM | chr17 | 37881427 | 37881427 | C | T | Silent | p.D843D | 1 |
| ERBB2 | BLCA | chr17 | 37884259 | 37884259 | G | C | Missense_Mutation | p.E1214Q | 1 |
| ERBB2 | KIRP | chr17 | 37876080 | 37876080 | A | C | Silent | p.R617R | 1 |
| ERBB2 | LUAD | chr17 | 37868201 | 37868201 | G | A | Missense_Mutation | p.V278M | 1 |
 Copy number variation (CNV) of ERBB2 Copy number variation (CNV) of ERBB2 * Click on the image to open the original image in a new window. |
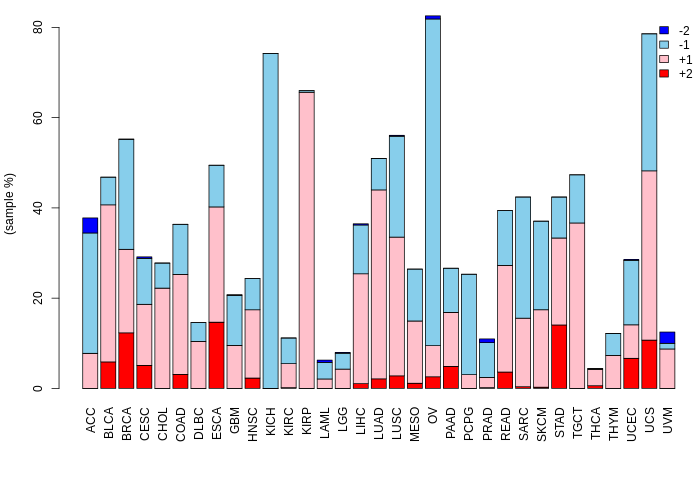 |
 Fusion gene breakpoints (product of the structural variants (SVs)) across ERBB2 Fusion gene breakpoints (product of the structural variants (SVs)) across ERBB2 * Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion genes with this translation factor from FusionGDB2.0. Fusion genes with this translation factor from FusionGDB2.0. |
| FusionGDB2 ID | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 101608 | N/A | AX187163 | ABCC5 | chr3 | 183716296 | - | ERBB2 | chr17 | 37870791 | + |
| 101608 | HNSC | TCGA-CV-5432-01A | ACACA | chr17 | 35472930 | - | ERBB2 | chr17 | 37863393 | - |
| 101608 | HNSC | TCGA-CV-5432-01A | ACACA | chr17 | 35479394 | - | ERBB2 | chr17 | 37863393 | - |
| 101608 | BRCA | TCGA-A8-A08X-01A | AP2B1 | chr17 | 33968994 | + | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-A8-A08X-01A | AP2B1 | chr17 | 33968994 | + | ERBB2 | chr17 | 37849514 | + |
| 101608 | N/A | DL055715 | ASH1L | chr1 | 155446335 | - | ERBB2 | chr17 | 37884815 | - |
| 101608 | STAD | TCGA-BR-4357 | CAB39 | chr2 | 231577945 | + | ERBB2 | chr17 | 37863242 | + |
| 101608 | BRCA | TCGA-D8-A1XJ-01A | CACNB1 | chr17 | 37334237 | - | ERBB2 | chr17 | 37886276 | + |
| 101608 | LUSC | TCGA-L3-A524-01A | CASC3 | chr17 | 38297860 | + | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-A8-A08X | CDK12 | chr17 | 37657692 | + | ERBB2 | chr17 | 37851240 | + |
| 101608 | BRCA | TCGA-A8-A08X-01A | CDK12 | chr17 | 37657692 | + | ERBB2 | chr17 | 37851241 | + |
| 101608 | BRCA | TCGA-C8-A12Q-01A | CORO1B | chr11 | 67207589 | - | ERBB2 | chr17 | 37868181 | + |
| 101608 | STAD | TCGA-CD-5799-01A | CPD | chr17 | 28706532 | + | ERBB2 | chr17 | 37875611 | + |
| 101608 | BRCA | TCGA-BH-A1F2-01A | EME1 | chr17 | 48452873 | + | ERBB2 | chr17 | 37882815 | + |
| 27204 | BRCA | TCGA-C8-A132-01A | ERBB2 | chr17 | 37881655 | + | ABI3 | chr17 | 47293893 | + |
| 27204 | BRCA | TCGA-C8-A132-01A | ERBB2 | chr17 | 37882106 | + | ABI3 | chr17 | 47293893 | + |
| 85929 | BRCA | TCGA-AO-A0JM-01A | ERBB2 | chr17 | 37868701 | + | ARL5C | chr17 | 37321392 | - |
| 100579 | N/A | BI045492 | ERBB2 | chr17 | 37874310 | - | CD99 | chrY | 2586265 | - |
| 99264 | BRCA | TCGA-BH-A204 | ERBB2 | chr17 | 37868701 | + | CDK12 | chr17 | 37680926 | + |
| 99264 | BRCA | TCGA-BH-A204-01A | ERBB2 | chr17 | 37868701 | + | CDK12 | chr17 | 37680927 | + |
| 99264 | BRCA | TCGA-E2-A1LE-01A | ERBB2 | chr17 | 37866734 | + | CDK12 | chr17 | 37712539 | + |
| 99264 | STAD | TCGA-BR-8289-01A | ERBB2 | chr17 | 37886762 | + | CDK12 | chr17 | 37667831 | + |
| 99264 | UCEC | TCGA-EO-A2CG-01A | ERBB2 | chr17 | 37873733 | + | CDK12 | chr17 | 37721013 | + |
| 82922 | BRCA | TCGA-UU-A93S-01A | ERBB2 | chr17 | 37884915 | - | CLDN7 | chr17 | 7166264 | - |
| 85705 | UCEC | TCGA-D1-A3JP-01A | ERBB2 | chr17 | 37863394 | + | CNIH4 | chr1 | 224553581 | + |
| 85705 | UCEC | TCGA-D1-A3JP-01A | ERBB2 | chr17 | 37866134 | + | CNIH4 | chr1 | 224553581 | + |
| 85705 | UCEC | TCGA-D1-A3JP-01A | ERBB2 | chr17 | 37866734 | + | CNIH4 | chr1 | 224553581 | + |
| 87284 | BLCA | TCGA-4Z-AA7S-01A | ERBB2 | chr17 | 37872192 | + | CTTN | chr11 | 70269046 | + |
| 86610 | COAD | TCGA-D5-6531-01A | ERBB2 | chr17 | 37844531 | + | DNAJC7 | chr17 | 40134419 | - |
| 93134 | STAD | TCGA-BR-A4PE | ERBB2 | chr17 | 37868300 | + | EIF1 | chr17 | 39846029 | + |
| 93134 | STAD | TCGA-BR-A4PE | ERBB2 | chr17 | 37868701 | + | EIF1 | chr17 | 39846029 | + |
| 93134 | STAD | TCGA-BR-A4PE-01A | ERBB2 | chr17 | 37868701 | + | EIF1 | chr17 | 39846030 | + |
| 78034 | STAD | TCGA-RD-A7BT-01A | ERBB2 | chr17 | 37856564 | + | FKBP10 | chr17 | 39974341 | + |
| 78034 | STAD | TCGA-RD-A7BT-01A | ERBB2 | chr17 | 37866734 | + | FKBP10 | chr17 | 39977906 | + |
| 102980 | STAD | TCGA-FP-8210-01A | ERBB2 | chr17 | 37872192 | + | GUK1 | chr1 | 228333727 | + |
| 101785 | BRCA | TCGA-D8-A1XJ-01A | ERBB2 | chr17 | 37882868 | + | HIAT1 | chr1 | 100534041 | + |
| 103189 | CESC | TCGA-C5-A2LX-01A | ERBB2 | chr17 | 37872858 | + | IKZF3 | chr17 | 37985741 | - |
| 103189 | ESCA | TCGA-R6-A8W5-01B | ERBB2 | chr17 | 37849578 | + | IKZF3 | chr17 | 37914218 | - |
| 103189 | HNSC | TCGA-CN-A49C-01A | ERBB2 | chr17 | 37873733 | + | IKZF3 | chr17 | 37949186 | - |
| 103189 | OV | TCGA-25-1320-01A | ERBB2 | chr17 | 37873733 | + | IKZF3 | chr17 | 37914218 | - |
| 103189 | READ | TCGA-EI-6509 | ERBB2 | chr17 | 37866134 | + | IKZF3 | chr17 | 37988404 | - |
| 103189 | STAD | TCGA-D7-8573-01A | ERBB2 | chr17 | 37886516 | + | IKZF3 | chr17 | 37949186 | - |
| 103189 | UCEC | TCGA-B5-A3S1-01A | ERBB2 | chr17 | 37879913 | + | IKZF3 | chr17 | 37944627 | - |
| 99272 | Non-Cancer | TCGA-V5-A7RE-11A | ERBB2 | chr17 | 37872192 | + | JUP | chr17 | 39911684 | - |
| 87242 | STAD | TCGA-BR-A4IZ | ERBB2 | chr17 | 37872192 | + | KAT2A | chr17 | 40267053 | - |
| 88945 | ESCA | TCGA-M9-A5M8 | ERBB2 | chr17 | 37851434 | + | KRT10 | chr17 | 38977369 | - |
| 88945 | ESCA | TCGA-M9-A5M8 | ERBB2 | chr17 | 37856564 | + | KRT10 | chr17 | 38977369 | - |
| 57254 | STAD | TCGA-CG-4466-01A | ERBB2 | chr17 | 37868701 | + | KRT24 | chr17 | 38858185 | - |
| 86250 | STAD | TCGA-BR-8483-01A | ERBB2 | chr17 | 37856564 | + | KRT39 | chr17 | 39116662 | - |
| 99417 | BRCA | TCGA-D8-A1XJ-01A | ERBB2 | chr17 | 37873733 | + | MED1 | chr17 | 37571592 | - |
| 97197 | STAD | TCGA-VQ-AA6G | ERBB2 | chr17 | 37876087 | + | MIEN1 | chr17 | 37886544 | - |
| 96452 | OV | TCGA-20-1686 | ERBB2 | chr17 | 37872192 | + | MUC1 | chr1 | 155159850 | - |
| 90848 | BRCA | TCGA-BH-A1EN | ERBB2 | chr17 | 37866134 | + | NARS2 | chr11 | 78147860 | - |
| 90848 | BRCA | TCGA-BH-A1EN | ERBB2 | chr17 | 37866734 | + | NARS2 | chr11 | 78147860 | - |
| 90848 | BRCA | TCGA-BH-A1EN-01A | ERBB2 | chr17 | 37866733 | + | NARS2 | chr11 | 78147859 | - |
| 71237 | CESC | TCGA-DS-A7WF | ERBB2 | chr17 | 37856564 | + | NEUROD2 | chr17 | 37762857 | - |
| 71237 | CESC | TCGA-DS-A7WF-01A | ERBB2 | chr17 | 37856563 | + | NEUROD2 | chr17 | 37762856 | - |
| 71237 | CESC | TCGA-VS-A9UL | ERBB2 | chr17 | 37851434 | + | NEUROD2 | chr17 | 37764318 | - |
| 71237 | CESC | TCGA-VS-A9UL | ERBB2 | chr17 | 37856564 | + | NEUROD2 | chr17 | 37764318 | - |
| 71237 | CESC | TCGA-VS-A9UL | ERBB2 | chr17 | 37866134 | + | NEUROD2 | chr17 | 37764318 | - |
| 100884 | BRCA | TCGA-C8-A137-01A | ERBB2 | chr17 | 37863394 | + | NFE2L2 | chr2 | 178098999 | - |
| 98627 | UCEC | TCGA-EY-A54A-01A | ERBB2 | chr17 | 37884523 | + | NOL12 | chr22 | 38077874 | + |
| 100918 | N/A | CV340327 | ERBB2 | chr17 | 37884125 | + | NOMO1 | chr16 | 14976494 | + |
| 27204 | STAD | TCGA-FP-8211-01A | ERBB2 | chr17 | 37872192 | + | NUDT8 | chr11 | 67396522 | - |
| 99077 | CESC | TCGA-Q1-A73R | ERBB2 | chr17 | 37851434 | + | PGAP3 | chr17 | 37830932 | - |
| 99077 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37851433 | + | PGAP3 | chr17 | 37830931 | - |
| 99077 | ESCA | TCGA-2H-A9GF | ERBB2 | chr17 | 37856564 | + | PGAP3 | chr17 | 37830932 | - |
| 99077 | ESCA | TCGA-2H-A9GF | ERBB2 | chr17 | 37856564 | + | PGAP3 | chr17 | 37841002 | - |
| 99077 | ESCA | TCGA-2H-A9GF | ERBB2 | chr17 | 37856564 | + | PGAP3 | chr17 | 37842272 | - |
| 99077 | UCEC | TCGA-FI-A2F8 | ERBB2 | chr17 | 37866734 | + | PGAP3 | chr17 | 37842272 | - |
| 89643 | CESC | TCGA-Q1-A73R | ERBB2 | chr17 | 37851434 | + | PNMT | chr17 | 37825881 | + |
| 103131 | BLCA | TCGA-DK-A1A3-01A | ERBB2 | chr17 | 37866734 | + | PPP1R1B | chr17 | 37790136 | + |
| 103131 | BRCA | TCGA-C8-A12Z-01A | ERBB2 | chr17 | 37872192 | + | PPP1R1B | chr17 | 37790136 | + |
| 103131 | CESC | TCGA-4J-AA1J-01A | ERBB2 | chr17 | 37856564 | + | PPP1R1B | chr17 | 37790136 | + |
| 103131 | CESC | TCGA-Q1-A73R | ERBB2 | chr17 | 37883256 | + | PPP1R1B | chr17 | 37791859 | + |
| 103131 | CESC | TCGA-Q1-A73R | ERBB2 | chr17 | 37883800 | + | PPP1R1B | chr17 | 37791859 | + |
| 103131 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37883255 | + | PPP1R1B | chr17 | 37791859 | + |
| 103131 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37883256 | + | PPP1R1B | chr17 | 37791860 | + |
| 103131 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37883799 | + | PPP1R1B | chr17 | 37791859 | + |
| 103131 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37883800 | + | PPP1R1B | chr17 | 37791860 | + |
| 103131 | LUSC | TCGA-22-0944-01A | ERBB2 | chr17 | 37876087 | + | PPP1R1B | chr17 | 37792069 | + |
| 103131 | OV | TCGA-24-2033-01A | ERBB2 | chr17 | 37856564 | + | PPP1R1B | chr17 | 37785423 | + |
| 103131 | STAD | TCGA-RD-A7BT-01A | ERBB2 | chr17 | 37886516 | + | PPP1R1B | chr17 | 37783559 | + |
| 86255 | BRCA | TCGA-A2-A1G1-01A | ERBB2 | chr17 | 37879913 | + | PSMB3 | chr17 | 36920372 | + |
| 86255 | ESCA | TCGA-JY-A6FA | ERBB2 | chr17 | 37876087 | + | PSMB3 | chr17 | 36916683 | + |
| 86255 | ESCA | TCGA-JY-A6FA-01A | ERBB2 | chr17 | 37876087 | + | PSMB3 | chr17 | 36916684 | + |
| 86255 | ESCA | TCGA-L5-A88V | ERBB2 | chr17 | 37873733 | + | PSMB3 | chr17 | 36916683 | + |
| 86255 | ESCA | TCGA-L5-A88V | ERBB2 | chr17 | 37876087 | + | PSMB3 | chr17 | 36918663 | + |
| 99282 | STAD | TCGA-BR-8059-01A | ERBB2 | chr17 | 37863394 | + | PSMD3 | chr17 | 38144936 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37873733 | + | PVT1 | chr8 | 129082406 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37873733 | + | PVT1 | chr8 | 129108764 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37873733 | + | PVT1 | chr8 | 129108767 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37876086 | + | PVT1 | chr8 | 129043188 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37876086 | + | PVT1 | chr8 | 129043282 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37876087 | + | PVT1 | chr8 | 129082406 | + |
| 102270 | UCEC | TCGA-EY-A3QX-01A | ERBB2 | chr17 | 37876087 | + | PVT1 | chr8 | 129108764 | + |
| 79988 | N/A | AW371253 | ERBB2 | chr17 | 37881986 | + | RABGAP1 | chr9 | 125852605 | + |
| 27204 | UCEC | TCGA-AP-A053-01A | ERBB2 | chr17 | 37881398 | + | RARRES2 | chr7 | 150037762 | - |
| 83930 | BRCA | TCGA-C8-A8HP-01A | ERBB2 | chr17 | 37856564 | + | SDF4 | chr1 | 1159348 | - |
| 100123 | BRCA | TCGA-AR-A254 | ERBB2 | chr17 | 37883256 | + | SLC29A3 | chr10 | 73121710 | + |
| 100123 | BRCA | TCGA-AR-A254-01A | ERBB2 | chr17 | 37883255 | + | SLC29A3 | chr10 | 73121710 | + |
| 100123 | BRCA | TCGA-AR-A254-01A | ERBB2 | chr17 | 37883256 | + | SLC29A3 | chr10 | 73121711 | + |
| 97282 | BRCA | TCGA-A2-A0YE-01A | ERBB2 | chr17 | 37871789 | - | SLC39A11 | chr17 | 71084914 | - |
| 93086 | BRCA | TCGA-A2-A3XZ-01A | ERBB2 | chr17 | 37884002 | + | SMARCE1 | chr17 | 38787057 | - |
| 85104 | BRCA | TCGA-BH-A1EV-01A | ERBB2 | chr17 | 37872192 | + | SRCIN1 | chr17 | 36699371 | - |
| 99287 | CESC | TCGA-Q1-A73R | ERBB2 | chr17 | 37866734 | + | STARD3 | chr17 | 37815303 | + |
| 99287 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37866733 | + | STARD3 | chr17 | 37815303 | + |
| 99287 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37872857 | + | STARD3 | chr17 | 37809733 | + |
| 99287 | CESC | TCGA-Q1-A73R-01A | ERBB2 | chr17 | 37873732 | + | STARD3 | chr17 | 37809733 | + |
| 99287 | ESCA | TCGA-M9-A5M8 | ERBB2 | chr17 | 37872858 | + | STARD3 | chr17 | 37816463 | + |
| 99287 | STAD | TCGA-CD-5799-01A | ERBB2 | chr17 | 37856564 | + | STARD3 | chr17 | 37809734 | + |
| 97624 | BRCA | TCGA-BH-A0DZ-11A | ERBB2 | chr17 | 37881654 | + | STXBP4 | chr17 | 53065831 | - |
| 97624 | BRCA | TCGA-BH-A0DZ-11A | ERBB2 | chr17 | 37881847 | + | STXBP4 | chr17 | 53065852 | - |
| 98320 | LUAD | TCGA-69-7761-01A | ERBB2 | chr17 | 37884915 | - | SUPT16H | chr14 | 21829491 | - |
| 92224 | BLCA | TCGA-DK-A2I6-01A | ERBB2 | chr17 | 37868701 | + | TATDN1 | chr8 | 125507783 | - |
| 76256 | LUSC | TCGA-56-A4BY | ERBB2 | chr17 | 37872858 | + | TCAP | chr17 | 37821626 | + |
| 76256 | LUSC | TCGA-56-A4BY | ERBB2 | chr17 | 37873451 | + | TCAP | chr17 | 37821131 | + |
| 76256 | LUSC | TCGA-56-A4BY-01A | ERBB2 | chr17 | 37872858 | + | TCAP | chr17 | 37821969 | + |
| 99290 | BRCA | TCGA-GM-A2DA-01A | ERBB2 | chr17 | 37880176 | + | THRA | chr17 | 38233860 | - |
| 99295 | STAD | TCGA-D7-8573 | ERBB2 | chr17 | 37873733 | + | WIPF2 | chr17 | 38412642 | + |
| 99295 | STAD | TCGA-D7-8573-01A | ERBB2 | chr17 | 37873733 | + | WIPF2 | chr17 | 38412643 | + |
| 99422 | STAD | TCGA-VQ-A8DZ-01A | ERBB2 | chr17 | 37884055 | + | WSB1 | chr17 | 25628814 | + |
| 99422 | STAD | TCGA-VQ-A8DZ-01A | ERBB2 | chr17 | 37884130 | + | WSB1 | chr17 | 25625630 | + |
| 99422 | STAD | TCGA-VQ-A8DZ-01A | ERBB2 | chr17 | 37884131 | + | WSB1 | chr17 | 25625631 | + |
| 102744 | BRCA | TCGA-OL-A5RY-01A | ERBB2 | chr17 | 37866734 | + | ZAN | chr7 | 100392818 | + |
| 99298 | STAD | TCGA-BR-A4PE | ERBB2 | chr17 | 37882106 | + | ZPBP2 | chr17 | 38026946 | + |
| 101608 | BRCA | TCGA-A8-A09I-01A | FBXL20 | chr17 | 37557614 | - | ERBB2 | chr17 | 37863243 | + |
| 101608 | STAD | TCGA-BR-8483-01A | FKBP10 | chr17 | 39969531 | + | ERBB2 | chr17 | 37855790 | + |
| 101608 | BRCA | TCGA-D8-A1X5-01A | GPR125 | chr4 | 22475395 | - | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-D8-A1X5-01A | GPR125 | chr4 | 22475395 | - | ERBB2 | chr17 | 37849514 | + |
| 101608 | BLCA | TCGA-GU-AATP | GRB7 | chr17 | 37894386 | + | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-C8-A1HK-01A | GRB7 | chr17 | 37902253 | + | ERBB2 | chr17 | 37883942 | + |
| 101608 | CESC | TCGA-C5-A1M9-01A | GRB7 | chr17 | 37902455 | + | ERBB2 | chr17 | 37849514 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37897160 | + | ERBB2 | chr17 | 37879791 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37898709 | + | ERBB2 | chr17 | 37855790 | + |
| 101608 | CESC | TCGA-LP-A5U3-01A | GRB7 | chr17 | 37898709 | + | ERBB2 | chr17 | 37863243 | + |
| 101608 | CESC | TCGA-VS-A9V5 | GRB7 | chr17 | 37894386 | + | ERBB2 | chr17 | 37886276 | + |
| 101608 | CESC | TCGA-VS-A9V5 | GRB7 | chr17 | 37894645 | + | ERBB2 | chr17 | 37886276 | + |
| 101608 | ESCA | TCGA-V5-A7RE | KIAA0100 | chr17 | 26958497 | - | ERBB2 | chr17 | 37849514 | + |
| 101608 | . | . | MDK | chr11 | 46404136 | + | ERBB2 | chr17 | 46404136 | + |
| 101608 | BRCA | TCGA-BH-A1EV | MED24 | chr17 | 38188901 | - | ERBB2 | chr17 | 37868574 | + |
| 101608 | STAD | TCGA-HU-A4G6 | MED24 | chr17 | 38185078 | - | ERBB2 | chr17 | 37865570 | + |
| 101608 | STAD | TCGA-EQ-8122 | MIB2 | chr1 | 1551994 | + | ERBB2 | chr17 | 37863242 | + |
| 101608 | CESC | TCGA-EK-A2RM-01A | MIEN1 | chr17 | 37885540 | - | ERBB2 | chr17 | 37863243 | + |
| 101608 | BLCA | TCGA-DK-A2I6 | MTSS1 | chr8 | 125740124 | - | ERBB2 | chr17 | 37863242 | + |
| 101608 | BLCA | TCGA-DK-A2I6-01A | MTSS1 | chr8 | 125740125 | - | ERBB2 | chr17 | 37863243 | + |
| 101608 | BRCA | TCGA-C8-A275-01A | MYO18A | chr17 | 27464034 | - | ERBB2 | chr17 | 37879572 | + |
| 101608 | BRCA | TCGA-C8-A275-01A | MYO18A | chr17 | 27492960 | - | ERBB2 | chr17 | 37879791 | + |
| 101608 | BLCA | TCGA-2F-A9KP | PGAP3 | chr17 | 37844086 | - | ERBB2 | chr17 | 37844948 | + |
| 101608 | BLCA | TCGA-2F-A9KP-01A | PGAP3 | chr17 | 37840850 | - | ERBB2 | chr17 | 37883942 | + |
| 101608 | CESC | TCGA-DS-A7WF-01A | PGAP3 | chr17 | 37837498 | - | ERBB2 | chr17 | 37883650 | - |
| 101608 | CESC | TCGA-VS-A9V0-01A | PGAP3 | chr17 | 37840850 | - | ERBB2 | chr17 | 37880979 | + |
| 101608 | ESCA | TCGA-L5-A8NJ | PGAP3 | chr17 | 37840849 | - | ERBB2 | chr17 | 37844948 | + |
| 101608 | ESCA | TCGA-L5-A8NJ | PGAP3 | chr17 | 37840850 | - | ERBB2 | chr17 | 37844949 | + |
| 101608 | ESCA | TCGA-L5-A8NJ | PGAP3 | chr17 | 37840850 | - | ERBB2 | chr17 | 37849514 | + |
| 101608 | STAD | TCGA-D7-6822-01A | PGAP3 | chr17 | 37842175 | - | ERBB2 | chr17 | 37844949 | + |
| 101608 | BRCA | TCGA-A8-A08X-01A | PIP4K2B | chr17 | 36955519 | - | ERBB2 | chr17 | 37855790 | + |
| 101608 | N/A | BE696969 | PLEC | chr8 | 145012593 | - | ERBB2 | chr17 | 37864712 | - |
| 101608 | BRCA | TCGA-D8-A1XJ-01A | PLXDC1 | chr17 | 37306531 | - | ERBB2 | chr17 | 37868700 | - |
| 101608 | N/A | BF837831 | PPFIA3 | chr19 | 49653673 | - | ERBB2 | chr17 | 37881146 | - |
| 101608 | BRCA | TCGA-UL-AAZ6-01A | PPP1R1B | chr17 | 37783927 | + | ERBB2 | chr17 | 37879912 | - |
| 101608 | BRCA | TCGA-UL-AAZ6-01A | PPP1R1B | chr17 | 37783962 | + | ERBB2 | chr17 | 37879912 | - |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36916703 | + | ERBB2 | chr17 | 37872634 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36916746 | + | ERBB2 | chr17 | 37873665 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36916751 | + | ERBB2 | chr17 | 37872131 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36918690 | + | ERBB2 | chr17 | 37866433 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36920430 | + | ERBB2 | chr17 | 37873594 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36920433 | + | ERBB2 | chr17 | 37872612 | + |
| 101608 | ESCA | TCGA-L5-A88V | PSMB3 | chr17 | 36920471 | + | ERBB2 | chr17 | 37871734 | + |
| 101608 | BRCA | TCGA-E2-A152-01A | SLC2A1 | chr1 | 43392014 | - | ERBB2 | chr17 | 37868586 | + |
| 101608 | LUAD | TCGA-44-A4SS-01A | ST14 | chr11 | 130059791 | + | ERBB2 | chr17 | 37873573 | + |
| 101608 | N/A | BF229957 | TMEM177 | chr2 | 120473042 | + | ERBB2 | chr17 | 37866403 | - |
| 101608 | STAD | TCGA-R5-A7ZE | WIPF2 | chr17 | 38421398 | + | ERBB2 | chr17 | 37872553 | + |
| 101608 | STAD | TCGA-R5-A7ZE-01B | WIPF2 | chr17 | 38421398 | + | ERBB2 | chr17 | 37872554 | + |
| 101620 | . | . | ZNF207 | chr17 | 30683717 | ERBB2 | chr17 | 37873369 |
Top |
|
 Kaplan-Meier plots with logrank tests of overall survival (OS) Kaplan-Meier plots with logrank tests of overall survival (OS) |
 |
| Cancer type | Translation factor | Coefficent | Hazard ratio | Wald test pval | Likelihool ratio pval | Logrank test pval | # samples |
Top |
|
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05) Differential gene expression between female and male. (Wilcoxon test, pval<0.05) |
 |
| Cancer type | Translation factor | pval | adj.p |
| STAD | ERBB2 | 0.000880929568306567 | 0.024 |
| TGCT | ERBB2 | 4.89135747722409e-05 | 0.0014 |
Top |
|
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05) Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05) |
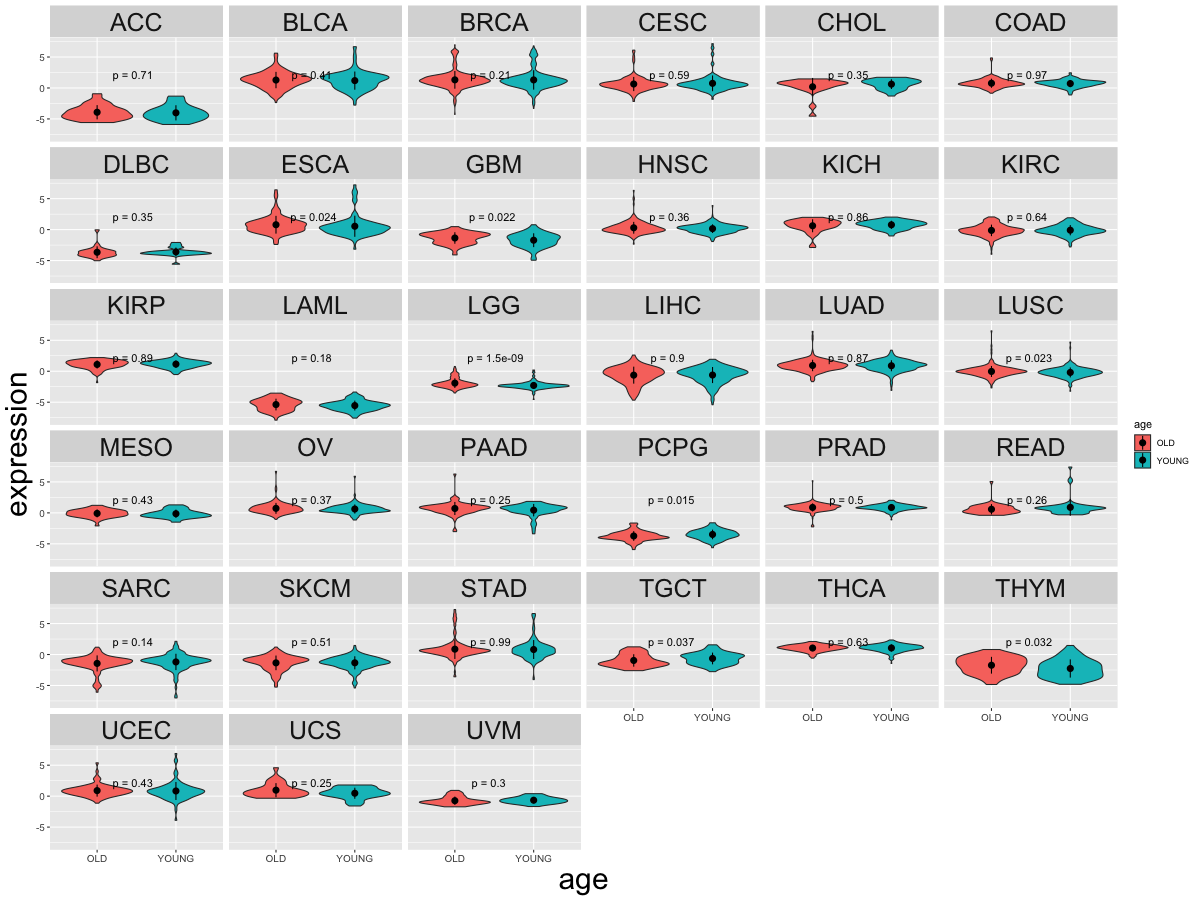 |
| Cancer type | Translation factor | pval | adj.p |
| TGCT | ERBB2 | 0.0369015224251232 | 1 |
| LUSC | ERBB2 | 0.022569126071267 | 0.68 |
| GBM | ERBB2 | 0.0218069978233243 | 0.68 |
| LGG | ERBB2 | 1.54296043607822e-09 | 5.1e-08 |
| ESCA | ERBB2 | 0.0240873694179042 | 0.7 |
| PCPG | ERBB2 | 0.0153555014158034 | 0.49 |
| THYM | ERBB2 | 0.0324822273713568 | 0.91 |
Top |
|
 Drugs targeting genes involved in this translation factor. Drugs targeting genes involved in this translation factor. (DrugBank Version 5.1.8 2021-05-08) |
| UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
| P04626 | DB00072 | Trastuzumab | Antibody | Biotech | Approved|Investigational |
| P04626 | DB01259 | Lapatinib | Antagonist | Small molecule | Approved|Investigational |
| P04626 | DB04988 | IGN311 | Biotech | Investigational | |
| P04626 | DB05773 | Trastuzumab emtansine | Antibody | Biotech | Approved|Investigational |
| P04626 | DB05944 | Varlitinib | Small molecule | Investigational | |
| P04626 | DB06021 | AV-412 | Small molecule | Investigational | |
| P04626 | DB06366 | Pertuzumab | Inhibitor | Biotech | Approved |
| P04626 | DB08916 | Afatinib | Inhibitor | Small molecule | Approved |
| P04626 | DB11652 | Tucatinib | Inhibitor | Small molecule | Approved|Investigational |
| P04626 | DB11973 | Tesevatinib | Small molecule | Investigational | |
| P04626 | DB12267 | Brigatinib | Inhibitor | Small molecule | Approved|Investigational |
| P04626 | DB14967 | Margetuximab | Antagonist | Biotech | Investigational |
| P04626 | DB15035 | Zanubrutinib | Inhibitor | Small molecule | Approved|Investigational |
| P04626 | DB00072 | Trastuzumab | Antibody | ||
| P04626 | DB01259 | Lapatinib | Antagonist | ||
| P04626 | DB04988 | IGN311 | |||
| P04626 | DB05773 | Trastuzumab emtansine | Antibody | ||
| P04626 | DB05944 | Varlitinib | |||
| P04626 | DB06021 | AV-412 | |||
| P04626 | DB06366 | Pertuzumab | Inhibitor | ||
| P04626 | DB08916 | Afatinib | Inhibitor | ||
| P04626 | DB11652 | Tucatinib | Inhibitor | ||
| P04626 | DB11973 | Tesevatinib | |||
| P04626 | DB12267 | Brigatinib | Inhibitor | ||
| P04626 | DB14967 | Margetuximab | Antagonist | ||
| P04626 | DB15035 | Zanubrutinib | Inhibitor |
Top |
|
 Diseases associated with this translation factor. Diseases associated with this translation factor. (DisGeNet 4.0) |
| Disease ID | Disease Name | # PubMeds | Disease source |
| C0006142 | Malignant neoplasm of breast | 40 | CGI;CTD_human |
| C0678222 | Breast Carcinoma | 40 | CGI;CTD_human |
| C1257931 | Mammary Neoplasms, Human | 40 | CTD_human |
| C4704874 | Mammary Carcinoma, Human | 40 | CTD_human |
| C1140680 | Malignant neoplasm of ovary | 4 | CGI;CTD_human;UNIPROT |
| C0024623 | Malignant neoplasm of stomach | 3 | CGI;CTD_human |
| C0376358 | Malignant neoplasm of prostate | 3 | CTD_human |
| C1708349 | Hereditary Diffuse Gastric Cancer | 3 | CTD_human |
| C0205641 | Adenocarcinoma, Basal Cell | 2 | CTD_human |
| C0205642 | Adenocarcinoma, Oxyphilic | 2 | CTD_human |
| C0205643 | Carcinoma, Cribriform | 2 | CTD_human |
| C0205644 | Carcinoma, Granular Cell | 2 | CTD_human |
| C0205645 | Adenocarcinoma, Tubular | 2 | CTD_human |
| C0699791 | Stomach Carcinoma | 2 | CGI;UNIPROT |
| C0919267 | ovarian neoplasm | 2 | CGI;CTD_human |
| C0007102 | Malignant tumor of colon | 1 | CTD_human |
| C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| C0013930 | Embolism, Tumor | 1 | CTD_human |
| C0016978 | gallbladder neoplasm | 1 | CTD_human |
| C0021367 | Mammary Ductal Carcinoma | 1 | CTD_human |
| C0024232 | Lymphatic Metastasis | 1 | CTD_human |
| C0025149 | Medulloblastoma | 1 | CTD_human |
| C0030354 | Papilloma | 1 | CTD_human |
| C0153452 | Malignant neoplasm of gallbladder | 1 | CTD_human |
| C0205833 | Medullomyoblastoma | 1 | CTD_human |
| C0205874 | Papilloma, Squamous Cell | 1 | CTD_human |
| C0205875 | Papillomatosis | 1 | CTD_human |
| C0206698 | Cholangiocarcinoma | 1 | CTD_human |
| C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| C0278510 | Childhood Medulloblastoma | 1 | CTD_human |
| C0278876 | Adult Medulloblastoma | 1 | CTD_human |
| C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| C0345905 | Intrahepatic Cholangiocarcinoma | 1 | CTD_human |
| C0546837 | Malignant neoplasm of esophagus | 1 | CTD_human |
| C0684249 | Carcinoma of lung | 1 | UNIPROT |
| C0685938 | Malignant neoplasm of gastrointestinal tract | 1 | CTD_human |
| C0751291 | Desmoplastic Medulloblastoma | 1 | CTD_human |
| C1134719 | Invasive Ductal Breast Carcinoma | 1 | CTD_human |
| C1257925 | Mammary Carcinoma, Animal | 1 | CTD_human |
| C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| C1275668 | Melanotic medulloblastoma | 1 | CTD_human |
| C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| C1368275 | Pigmented Basal Cell Carcinoma | 1 | CTD_human |
| C2931822 | Nasopharyngeal carcinoma | 1 | CTD_human |
| C3805278 | Extrahepatic Cholangiocarcinoma | 1 | CTD_human |
| C4721610 | Carcinoma, Ovarian Epithelial | 1 | CTD_human |
| C4721806 | Carcinoma, Basal Cell | 1 | CTD_human |

