| Gene | Chr | Position | RefSeq | VarSeq | RefSeeq | VarType | Pathogenic | Disease | VarInfo |
| EEF1A2 | chr20 | 62119654 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119659 | CCGCCTTCTG | C | Microsatellite | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| EEF1A2 | chr20 | 62119660 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119661 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119669 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119698 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119702 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119705 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119708 | G | A | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119708 | G | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119717 | C | T | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119723 | G | A | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119734 | C | A | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119734 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119741 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119741 | G | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119747 | C | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119747 | C | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119747 | C | T | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119748 | G | A | single_nucleotide_variant | Likely_pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119754 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119762 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119768 | G | T | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119774 | G | A | single_nucleotide_variant | Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119776 | G | A | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Epileptic_encephalopathy,_early_infantile,_33|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62119777 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119777 | G | GC | Duplication | Likely_pathogenic | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| EEF1A2 | chr20 | 62119777 | G | T | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62119780 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001574|splice_acceptor_variant | SO:0001574|splice_acceptor_variant |
| EEF1A2 | chr20 | 62119782 | G | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119786 | G | C | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119787 | C | CG | Duplication | Benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119787 | CG | C | Deletion | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119793 | G | A | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119813 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119822 | A | AC | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119822 | AC | A | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62119875 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120030 | C | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120106 | A | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120261 | C | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120265 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120272 | G | A | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120302 | A | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120317 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120320 | C | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120332 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120332 | G | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120339 | G | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120353 | C | A | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120370 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120376 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120385 | C | G | single_nucleotide_variant | Likely_pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120394 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Intellectual_disability|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120397 | C | G | single_nucleotide_variant | Likely_pathogenic | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120398 | A | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120399 | A | G | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120403 | TCTC | T | Deletion | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001822|inframe_deletion | SO:0001822|inframe_deletion |
| EEF1A2 | chr20 | 62120433 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120440 | T | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120470 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120471 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62120485 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62120510 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120513 | C | T | single_nucleotide_variant | Benign | Intellectual_disability|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120515 | G | T | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120535 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120567 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120569 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120639 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120725 | A | AAGGGGC | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62120773 | C | CG | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121665 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121702 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121726 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121801 | C | CCAGCCCCCTGGACCCAGCG | Duplication | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121819 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121820 | G | A | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121821 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121826 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121828 | C | G | single_nucleotide_variant | Benign/Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121829 | C | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121829 | C | G | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62121841 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121849 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121859 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121862 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121877 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121890 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121900 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121903 | T | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121907 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121913 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121918 | C | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121919 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121928 | A | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121931 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121937 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121940 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121946 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121949 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121961 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121966 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121973 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121973 | G | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121985 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121988 | T | C | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62121998 | T | A | single_nucleotide_variant | Uncertain_significance | Inborn_genetic_diseases | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62121998 | T | C | single_nucleotide_variant | Likely_pathogenic | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122003 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122018 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122019 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122040 | G | A | single_nucleotide_variant | Likely_pathogenic | Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122054 | G | A | single_nucleotide_variant | Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122065 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|EEF1A2-related_disorders|Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122069 | C | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122072 | G | A | single_nucleotide_variant | Benign/Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122078 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62122079 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122082 | C | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62122094 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122095 | A | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122103 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122211 | G | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122230 | A | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122257 | AG | A | Deletion | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122286 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62122290 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124168 | A | AGGAT | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124182 | A | AATGG | Microsatellite | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124459 | G | T | single_nucleotide_variant | Benign | Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124477 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124480 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124490 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124491 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124498 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124503 | C | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124508 | C | G | single_nucleotide_variant | Pathogenic | Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124515 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124523 | G | A | single_nucleotide_variant | Uncertain_significance | Seizures|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124524 | C | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124527 | G | C | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124536 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124543 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124545 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124546 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124548 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124548 | G | C | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124548 | G | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124550 | G | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124569 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124580 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124587 | G | A | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124589 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124590 | G | A | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124596 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124616 | C | T | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124624 | C | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124629 | G | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62124636 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62124644 | G | C | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124666 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124674 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62124731 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62125890 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62125933 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62125934 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62125999 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126148 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126149 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126158 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126159 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126185 | A | G | single_nucleotide_variant | Benign | Seizures|Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126188 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126208 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126218 | G | A | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126236 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126236 | G | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126239 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126243 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126247 | T | A | single_nucleotide_variant | Uncertain_significance | Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126249 | T | G | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126253 | C | T | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126255 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126266 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126266 | G | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126273 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126274 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126275 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126278 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126289 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126290 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126292 | T | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126298 | CC | TT | Indel | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126299 | C | T | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126302 | C | T | single_nucleotide_variant | Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126308 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126314 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126320 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126327 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126332 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126353 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126354 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126355 | T | C | single_nucleotide_variant | Uncertain_significance | Seizures | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126367 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126379 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126380 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126381 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126403 | C | G | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126404 | C | T | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126405 | G | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126409 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | Intellectual_disability|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126410 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126415 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126416 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126422 | G | A | single_nucleotide_variant | Benign/Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126425 | C | T | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126428 | C | T | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126429 | G | A | single_nucleotide_variant | Uncertain_significance | Seizures | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62126434 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126437 | C | G | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126443 | T | C | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62126462 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126487 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126488 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126685 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62126745 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127116 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127121 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127203 | G | GC | Duplication | Conflicting_interpretations_of_pathogenicity | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127206 | C | T | single_nucleotide_variant | Likely_benign | Seizures | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127222 | G | A | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127228 | A | G | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127240 | A | C | single_nucleotide_variant | Likely_pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127245 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127248 | G | T | single_nucleotide_variant | Likely_pathogenic | EEF1A2-related_disorders | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127254 | G | A | single_nucleotide_variant | Benign/Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127257 | G | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127259 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127260 | A | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127262 | C | T | single_nucleotide_variant | Pathogenic/Likely_pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127278 | G | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001587|nonsense | SO:0001587|nonsense |
| EEF1A2 | chr20 | 62127293 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127296 | C | T | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127302 | G | C | single_nucleotide_variant | Likely_benign | not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127314 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127325 | C | T | single_nucleotide_variant | Pathogenic | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|Intellectual_disability|Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|Inborn_genetic_diseases|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127326 | G | A | single_nucleotide_variant | Benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127328 | G | A | single_nucleotide_variant | Likely_pathogenic | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127339 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127350 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127353 | C | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127353 | C | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127362 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127381 | T | C | single_nucleotide_variant | Uncertain_significance | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62127383 | C | A | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127383 | C | G | single_nucleotide_variant | Likely_benign | Seizures|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62127393 | AGC | A | Deletion | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127395 | C | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127395 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127396 | G | C | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127411 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127426 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127521 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127584 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62127618 | C | T | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62128735 | T | C | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62128895 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62128927 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62128946 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62128979 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62128980 | G | A | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62128988 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62128991 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_specified | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62128993 | A | T | single_nucleotide_variant | Likely_pathogenic | not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62128997 | C | T | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129000 | A | G | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129004 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129045 | C | T | single_nucleotide_variant | Benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129046 | G | A | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | Mental_retardation,_autosomal_dominant_38|Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129050 | T | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129063 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129063 | G | C | single_nucleotide_variant | Benign/Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129068 | C | G | single_nucleotide_variant | Conflicting_interpretations_of_pathogenicity | EEF1A2-related_developmental_and_degenerative_epileptic-dyskinetic_encephalopathy|Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129071 | C | G | single_nucleotide_variant | Likely_pathogenic | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129072 | G | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129077 | C | T | single_nucleotide_variant | Uncertain_significance | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129087 | G | A | single_nucleotide_variant | Benign/Likely_benign | Epileptic_encephalopathy,_early_infantile,_33|not_provided | SO:0001819|synonymous_variant | SO:0001819|synonymous_variant |
| EEF1A2 | chr20 | 62129089 | T | A | single_nucleotide_variant | Likely_benign | Epileptic_encephalopathy,_early_infantile,_33 | SO:0001583|missense_variant | SO:0001583|missense_variant |
| EEF1A2 | chr20 | 62129090 | GTTGATGTGGGTCT | G | Deletion | Uncertain_significance | not_provided | SO:0001589|frameshift_variant | SO:0001589|frameshift_variant |
| EEF1A2 | chr20 | 62129141 | C | A | single_nucleotide_variant | Likely_benign | not_specified | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| EEF1A2 | chr20 | 62129141 | C | T | single_nucleotide_variant | Benign | not_specified | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| EEF1A2 | chr20 | 62129148 | C | T | single_nucleotide_variant | Likely_benign | not_provided | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| EEF1A2 | chr20 | 62129149 | G | A | single_nucleotide_variant | Likely_benign | not_provided | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| EEF1A2 | chr20 | 62129168 | C | T | single_nucleotide_variant | Likely_benign | not_specified | SO:0001623|5_prime_UTR_variant | SO:0001623|5_prime_UTR_variant |
| EEF1A2 | chr20 | 62130311 | T | G | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62130351 | G | A | single_nucleotide_variant | Benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62130393 | GG | T | Indel | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62130403 | G | T | single_nucleotide_variant | Benign | not_specified | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62130404 | T | G | single_nucleotide_variant | Likely_benign | not_provided | SO:0001627|intron_variant | SO:0001627|intron_variant |
| EEF1A2 | chr20 | 62130469 | C | T | single_nucleotide_variant | Benign | not_provided | | |

 Gene Summary
Gene Summary Child GO biological process term(s) under GO:0006412
Child GO biological process term(s) under GO:0006412  Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez
Gene ontology of translaction factor with evidence of Inferred from Direct Assay (IDA) from Entrez  Inferred gene age of translation factor.
Inferred gene age of translation factor.  We searched PubMed using 'EEF1A2[title] AND translation [title] AND human.'
We searched PubMed using 'EEF1A2[title] AND translation [title] AND human.' Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure.
Skipped exons in TCGA and GTEx based on Ensembl gene isoform structure. 
 Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.
Open reading frame (ORF) analsis of exon skipping events based on Ensembl gene isoform structure.  Exon skipping position in the amino acid sequence.
Exon skipping position in the amino acid sequence. Potentially (partially) lost protein functional features of UniProt.
Potentially (partially) lost protein functional features of UniProt. Gene expression level across TCGA pancancer
Gene expression level across TCGA pancancer
 Gene expression level across GTEx pantissue
Gene expression level across GTEx pantissue
 Expression level of gene isoforms across TCGA pancancer
Expression level of gene isoforms across TCGA pancancer
 Expression level of gene isoforms across GTEx pantissue
Expression level of gene isoforms across GTEx pantissue
 Cancer(tissue) type-specific expression level of Translation factor using z-score distriution
Cancer(tissue) type-specific expression level of Translation factor using z-score distriution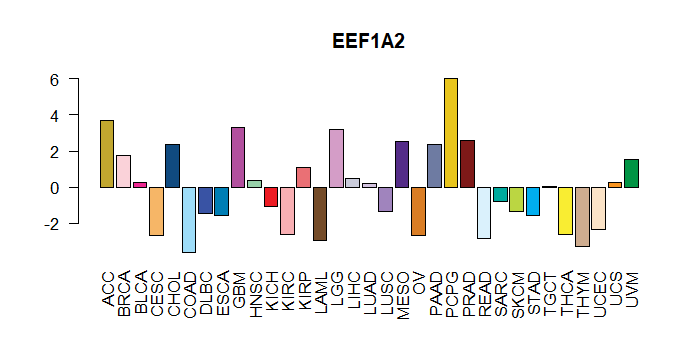
 Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)
Differential expression between tumor and matched normal (in the cancer types with more than 10 matched samples)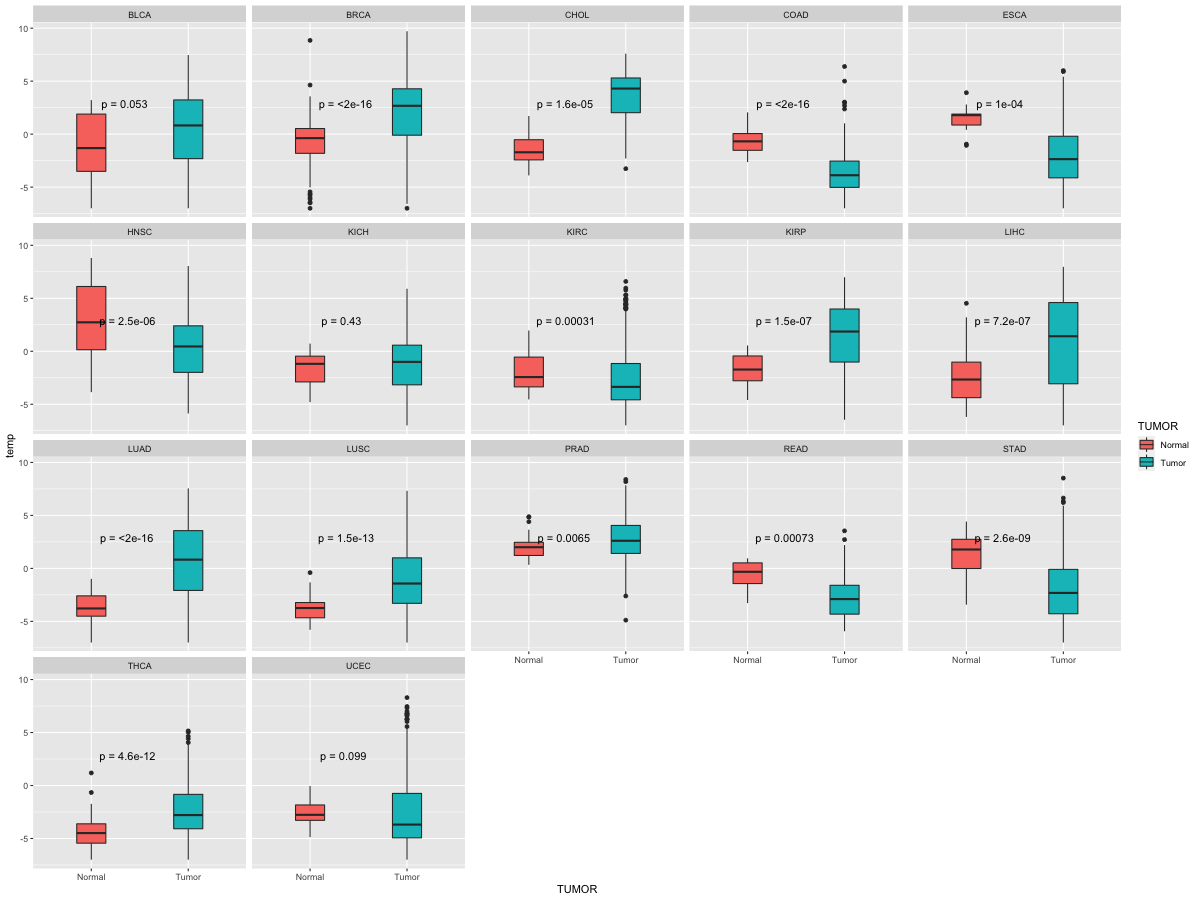
 Translation factor expression regulation through miRNA binding
Translation factor expression regulation through miRNA binding Translation factor expression regulation through methylation in the promoter of Translation factor
Translation factor expression regulation through methylation in the promoter of Translation factor
 Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
Translation factor expression regulation through methylation in the gene body of Translation factor (positive regulation)
 Translation factor expression regulation through copy number variation of Translation factor
Translation factor expression regulation through copy number variation of Translation factor 
 Strongly correlated genes belong to cellular important gene groups with EEF1A2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
Strongly correlated genes belong to cellular important gene groups with EEF1A2 (coefficient>0.8, pval<0.05, node color based on FC between tumor and matched normal). Significantly associated important genes in the individual cancer types. * Cell metabolism gene: cell metabolism genes from REACTOME (black edge), IUPHAR: drug target genes from IUPHAR (blue edge), Kinase: human kinase genes (brown edge), CGC: cancer gene census genes (orange edge), TSG: tumor suppresor genes (purple edge), Epifactor: epigenetic factors (light blue edge), TF: transcription factors (green)
 Protein 3D structure
Protein 3D structure Protein-protein interaction networks
Protein-protein interaction networks 
 Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))
Overlap between down-regulated DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network (center: Translation factor, node: DEGs, edges: weighted by -log2(adj.P))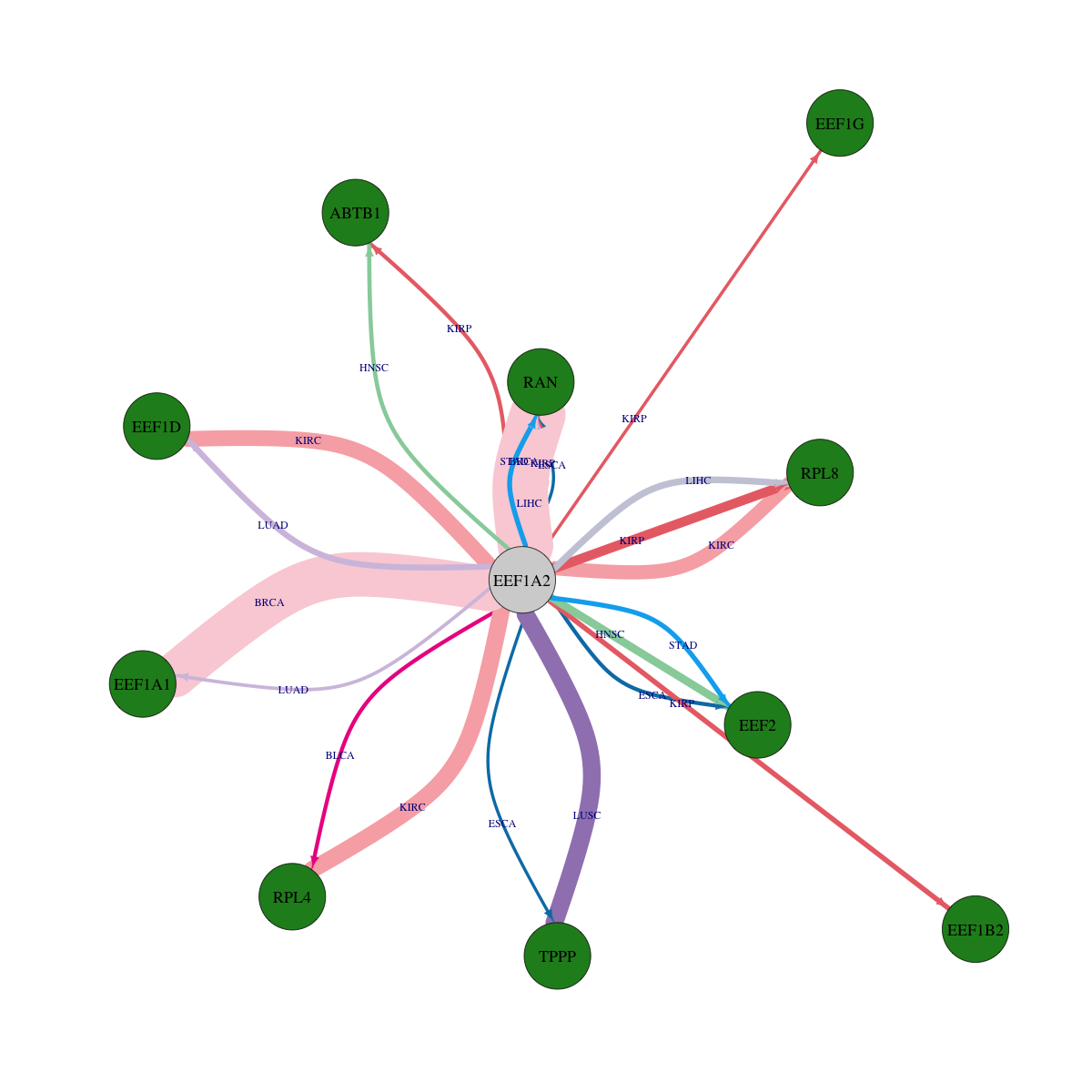
 * Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
* Overlap between DEGs (log2FC>1 and adj.P<0.05) and STRING PPI network per cancer (center: Translation factor, node: DEGs, node color: log2FC, edges: weighted by -log2(adj.P))
 Protein-protein interactors with this translation factor (BIOGRID-3.4.160)
Protein-protein interactors with this translation factor (BIOGRID-3.4.160) Clinically associated variants from ClinVar.
Clinically associated variants from ClinVar. nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.
nsSNVs with sample frequency (size of circle) from TCGA 33 cancers.  SNVs and Indels
SNVs and Indels Copy number variation (CNV) of EEF1A2
Copy number variation (CNV) of EEF1A2 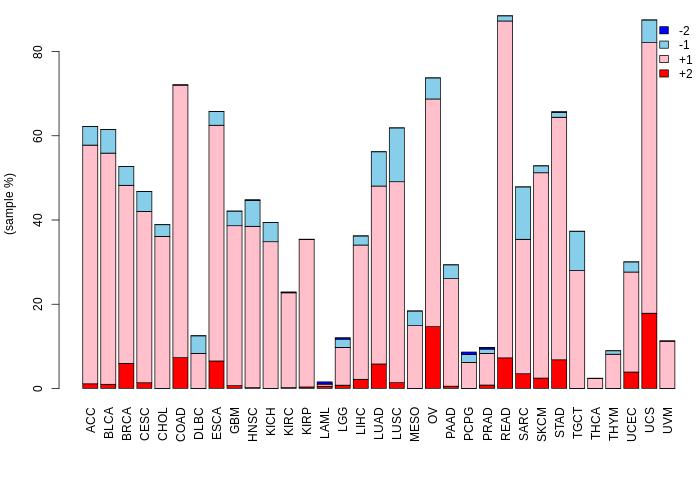
 Fusion gene breakpoints (product of the structural variants (SVs)) across EEF1A2
Fusion gene breakpoints (product of the structural variants (SVs)) across EEF1A2 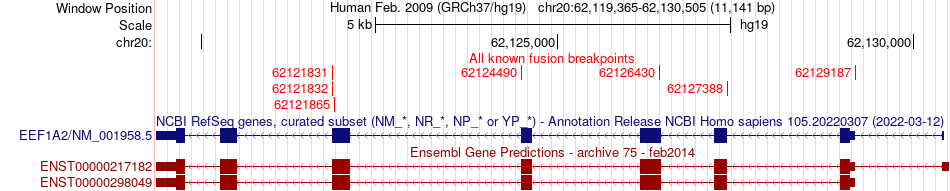
 Fusion genes with this translation factor from FusionGDB2.0.
Fusion genes with this translation factor from FusionGDB2.0.  Kaplan-Meier plots with logrank tests of overall survival (OS)
Kaplan-Meier plots with logrank tests of overall survival (OS) 
 Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
Differential gene expression between female and male. (Wilcoxon test, pval<0.05)
 Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
Differential gene expression between young and old age groups (Wilcoxon test, pval<0.05)
 Drugs targeting genes involved in this translation factor.
Drugs targeting genes involved in this translation factor.  Diseases associated with this translation factor.
Diseases associated with this translation factor. 
